Authors: Rajdeep Mundiyara1, Prem Kumar2 and Mamta Bajya3
1Seed Officer, Rajasthan State Seeds Corporation,Mandore, Jodhpure
2 Department of Plant Philology, Jobner
Email of corresponding author: rmundiyara5@gmail.com
RNA interference (RNAi) is a naturally occurring mechanism that leads to the “silencing” of genes. In consequence, the respective protein is no longer synthesised. In nature, this mechanism is used for the regulation of specific genes and is also applied as a defence against viruses. RNA interference (RNAi) is a form of post transcriptional gene regulation in which non translated double stranded RNA (dsRNA) molecules called small interfering RNA (siRNA) mediate sequence specific degradation of target messenger RNA (mRNA). RNA silencing is a novel gene regulatory mechanism that limits the transcript level by either suppressing transcription (TGS) or by activating a sequence- Specific RNA degradation process [PTGS/RNA interference (RNAi)]. The silencing effect was first observed in plants in 1990, when the Jorgensen laboratory introduced exogenous transgenes into petunias in an attempt to up-regulate the activity of a gene for chalcone synthase, an enzyme involved in the production of specific pigments (Agrawal et al., 2003). The natural function of RNAi is referring to the mechanism involved in cellular defense against viruses, genomic containment of retro-transposons, and post-transcriptional regulation of gene expression. RNAi can specifically silence individual genes, creating knockout phenotypes, either in transformants that can produce the required hairpin RNAs, or upon infection with recombinant RNA viruses that carry the target gene (VIGS, viral-induced gene silencing) (Tenea, 2009).RNAi is a multistep process involving the generation of small interfering RNAs (siRNAs) in vivo through the action of the RNase III endonuclease ‘Dicer’. The resulting 21- to 23-nt siRNAs mediate degradation of their complementary RNA (Zou et al., 2005). Recently two RNAi based crops have been given regulatory approval for commercial production and sale. These are the non-browning Arctic apples and the non-browning Innate potatoes. The firms producing these crops claim that the idea behind producing the non-browning apples and potatoes is not only to improve the look of the product, but it is also intended to increase the consumption of the raw fruits along with reducing naturally occurring carcinogens (as in the case of innate potatoes). While the science behind both these products is a little complicated as both are RNAi based, in simple way it can be put as both apples and potatoes have certain genes suppressed. Both of them, though genetically modified, are grown the same way as conventional varieties. These products are likely to find a place of attraction in the fresh-cut product sales.
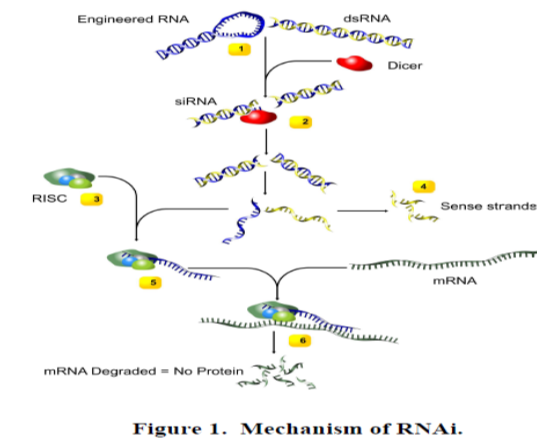
Mechanism of RNAi:
1. The entry of long double stranded RNA, such as an introduced transgene, a rogue genetic element or a viral intruder, triggers the RNAi pathway of cells. This results in the recruitment of the enzyme Dicer.
2. Dicer cleaves the dsRNA into short, 20-25 basepairs long, fragments, called small interfering RNA (siRNA).
3. An RNAinduced silencing complex (RISC) then distinguishes between the two siRNA strands as either sense or antisense. The sense strands (with exactly the same sequence as the target gene) are degraded.
4. The antisense strands on the other hand are incorporated to the RISC. These are used as guide to target messenger RNAs (mRNA) in a sequence specific manner.
5. Messenger RNAs (mRNA), which codes for amino acids, are cleaved by RISC. The activated RISC can repeatedly participate in mRNA degradation, inhibiting protein synthesis.
Application of RNAi technology
1. In plant system, it provides defense mechanism to protect against infection by viruses, transposons and other insertional elements.
2. RNAi also plays a role in regulating development and genome maintenance.
3. Development of male sterile plants in rice.
4. Application in improvement of nutritional value:
• RNAi technology used to produce cotton seed containing lower level of dcadinene
synthase which is key enzyme in gossypol production.
• RNAi method were used in cotton to down regulate two key fatty acid desaturase gene encoding stearoyl acyl careeer protein D9 desaturases and Oleoyl phasphatidylcholine w6 desaturase. Knockdown of these genes in cotton led to increase of nutritionally improved high oleic (HO) and high stearic (HS) cottonseed oil that is more suitable for human consumption.
• In maize, RNAi technology has been used to reduce phytic acid by silencing MRP4 ATP binding cassette (ABC) transporter.
• In soybean, Silencing of Omega3 fatty acid desaturase gene in soybean using RNAi reduce alinolenic acid and improve oil stability and flavour.
• Using RNAi technique, varieties of barley developed which are resistant to BYDV (barley yellow dwarf virus).
Advantages of RNAi
• This technology is highly gene specific.
• High gene silencing efficiency.
• Screening targeted plants takes less time.
• It is highly inducible.
Disadvantages of RNAi
• It does not knockout a gene for 100%.
• siRNA tends to activate unwanted pathways.
References:
Agrawal N, Dasaradhi P V N, Mohmmed A, Malhotra P, Bhatnagar R K, Mukherjee S K (2003). RNA Interference: Biology, Mechanism, and Applications. Microbiology and Molecular Biology Reviews 67: 657-685.
Tenea G N (2009). Exploring the world of RNA interference in plant functional genomics: a research tool for many biology phenomena, Centre of Microbial Biotechnology pp. 4360-4364.
Zou G M, Yoder Mervin C (2005). Application of RNA interference to study stem cell function: current status and future perspectives. Biol. Cell 97: 211�"219.
About Author / Additional Info: