Authors: Rakesh Kumar1,2, Amandeep Kaur1,2, H.M. Mamrutha1 and Pradeep Sharma1,*
1Crop Improvement Division, Directorate of Wheat Research, Karnal-132001, Haryana, India
2Department of Biotechnology Engineering, University Institute of Engineering & Technology, Kurukshetra University, Kurukshetra-136119, Haryana, India
*corresponding author: Email- neprads@yahoo.com (Pradeep Sharma)
Wheat is the second largest growing food cereal crop worldwide. It covers about 30% food requirement of world population. India has the second largest population after China. It is estimated that by 2030, India needs to produce 130 million tonnes of wheat to feed constantly growing population. This is the major challenge under variable climatic scenario in front of India as well as whole world. Although, all time high 95.91 million tonnes of wheat production recorded from an area of about 31 million hectare during 2013-2014 (First Advance Estimates 2014-15, Directorate of Economics and Statistics, Government of India). So we need to make more efforts to improve the wheat production under climatic variation to solve problems like abiotic stress (drought, heat, salt, alkalinity/sodicity) and biotic stress (rusts, leaf blight, powdery mildew) which highly reduce the production of wheat. Traditionally, we are improving wheat varieties using classical breeding approaches; consequently improvement has not happened with the pace of our requirement. We need to move towards modern breeding methods such as marker assisted selection and genetic engineering vis-a-vis over expression/RNAi methodologies to increase wheat quality and production. We have to generate a wheat variety that can bear wider range of changing climate variation with increase in yield. Before manipulating metabolic pathways of wheat or inserting a suitable gene or genes into the wheat, we should have the prior complete information of wheat genome which will tells us about the relationship among genes\metabolites present in the wheat. But this field has also a major limitation due to the largest and complex size of the wheat genome (Triticum aestivum L.; ~17 Gbp). Bread wheat is allohexaploid consisting of three closely-related sub genomes (A, B, D) and independently maintains genomes that are the result of a series of naturally occurring hybridization events. Each sub-genomes (each having 7 chromosomes i.e. 21 chromosome pairs) has almost 5500 million bases which are twice than human genome. Several individual groups around the world are working on sequencing of the wheat genome. Recently, the international wheat genome sequencing consortium (IWGSC) presented the draft sequence of 17 giga base pairs hexaploid bread wheat (T. aestivum cultivar ‘Chinese spring’) genome on 18th July, 2014. IWGSC annotated 124,201 gene loci well distributed evenly across the homologous chromosomes of sub-genomes. They used flow cytometer to sort individual chromosomes and generated pair-end sequence libraries with target of 500 base pair. Further, these libraries were sequenced using next generation sequence (NGS) technologies such as Roshce 454 and Illumina Sequencing technique. Using de novo gsAssembler (Newbler package) low copy-number genome assembly (LCG) were designed by removing repetitive sequences and by assembling remaining low-copy sequences. Mayers et al (2014) have provided a clear picture of bread wheat by comparing the genome of wheat with its wild relatives, such as tetraploid wheat cultivars (AABB) T. turgidum ‘Cappelli’ (originating from Italy) and T. turgidum 'Strongfield' (originating from Canada) and five diploid cultivars Aegilops speltoides (SS), T. urartu (AA), Ae. tauschii (DD), T. monococcum (AA), and Ae. sharonensis (SS) which provide the clue of bread wheat evolutionary history. It comes out that the T. urartu (the A-genome donor) and an unknown grass thought to be related to Ae. speltoides (the B-genome donor) are the ancestral progenitor genomes. This is considered that the first hybridization event produced tetraploid emmer wheat (AABB, T. dicoccoides) which hybridized again with Ae. tauschii (the D-genome donor) to produce modern bread wheat. The comparative gene analysis of wheat sub-genomes and its relative species genome showed the high similarity in respect of sequence and structural conservation. In another study, chromosome 3B was fully sequenced using a hybrid sequencing and BAC pooling strategy also known as hierarchical shotgun sequencing or a clone-by-clone shotgun strategy) (it is a rapid, cost effective method for genome). Chromosome 3B of bread wheat is largest with approximately 1-gigabase among other chromosomes consisting of 5326 protein coding genes, 1938 pseudogenes and 85% transposable elements. They also performed a comparative analysis which indicate that high wheat specific inter and intra-chromosomal gene duplication activities that are major source of variability for adaption. The IWGSC soon present the complete wheat genome which will helps in solving different mysteries present in wheat due to its huge genome. This will helps in understanding of different pathways of metabolism and roles of large number of genes in metabolic regulation. This chromosomes based genome sequence gives new insight into the genome size, structure, organisation and evolution of modern wheat.
A number of different applications can be fulfilled as the wheat genome comes:
• Development of novel molecular markers like SNPs, which helps in presentation of high resolution mapping of agronomically desirable traits, identifying candidate genes within a region of interest. These indentify traits then could be characterized and bred into elite wheat varieties.
• Functional and structural genomics: The genes can be characterised in respect of function and structure which enhance the trancriptomic and proteomic studies that permit deep insight into plant biology.
• Novel genes: A number of novel genes which are associated with valuable traits can be characterised from wheat, which can be transferred from wheat to wheat or to other species, from view of crop improvement, to make them more robust in nature.
• Allele mining: Genome sequence data would expedite sequence-enabled allele mining and accentuate it as an important pre-breeding tool for wheat improvement programs. It enables to determining allelic variation at candidate genes controlling key agronomic traits, identifying the evolution of alleles, predicting novel useful haplotypes, development of allele-specific markers for use in marker-assisted selection in wheat breeding program.
• Reference genome sequence availability will expedite: resequencing the genomes of any wheat cultivars, aligning with the reference genome for discovery of novel SNPs, SNPs assist in constructing high-resolution genetic maps, analysing the distribution of recombination, diversity along wheat chromosomes and to predict the new insertions of duplications or loss of genes. More clearly evolutionary history of wheat will come by comparing with relative species genome.
• High-throughput GBS approach would also serve as an option for efficient marker assisted breeding, genomic selection, and management of genetic resources at a higher ratio.
• miRNA and their regulation: NGS data can be investigated to predict miRNAs and their roles in gene regulation which can be used in improving stress tolerance, productivity of wheat cultivars and other agronomical traits.
Finally the wheat genome sequence will pave a fruitful pathway toward crop improvement, in producing the desired wheat genotype having capabilities for changing climate and helps to meet the challenge of sustaining our food supply in the coming years.
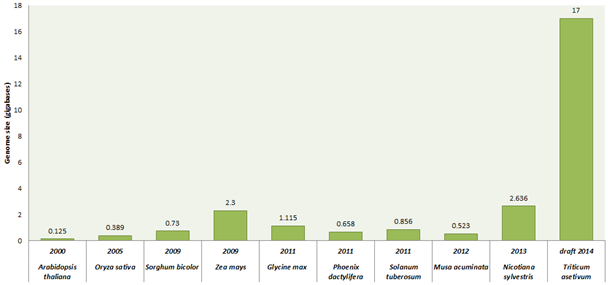
Figure 1: Genome sequencing projects of some important plants along with their year of completion and genome size.
About Author / Additional Info: