The function of proteins will be affected by the post translational modifications. Glycosylation is one such modification that supplements the protein structure with various oligo-saccharides. Glycosylation will diversify the protein and is also found to be involved in the activities like protein folding, receptor-ligand interaction, cell structure maintenance, cell-cell recognition and cell signaling. N-linked glycosylation is a co-translational process and post translational modification by which carbohydrates are attached to the asparagine at the region of consensus motif asparagine-X-serine/threonine. Here, X indicates any amino acid except proline. The motif NXC also attaches to glycan less frequently. Glycan attaches to the beta-amide group of asparagine. The beta-amide is a hydrogen bond donor which is donated to the oxygen of serine or threonine. Glycosylation process is catalyzed by N-glycosyltransferases which attach glycan to the protein that has not folded yet. It is proved experimentally that the primary structure of tri-peptide NXS/T is not enough for glycosylation to occur.
The evolution of glycosylated protein is found to occur slowly than the non-glycosylated protein of the same protein group. The structural restrictions of N-glycosylation can be developed as basic rules that improve the accuracy of the prediction. These rules are used to predict N-glycosylation of NXS/T consensus present in the human genome by single nucleotide variation. These rules can also be utilized in N-linked glycosylation prediction tool called as Sequence structure feature analysis tool (SFAT).
Results of the study
Structural analysis of NXS/T motif
Secondary structure conformation distribution in eukaryotes- Analysis of five eukaryotic proteomes of the mouse, plant, human, yeast and fly is performed in this study. The researchers worked on establishing the distribution of alpha-helix, beta-sheet and loop/turn in the available protein structures. The structural analysis revealed that all these structural elements are found to be similar in all the five organisms. But, the secondary structure conformations like alpha-helices were higher in number while beta-sheets were lowest. The frequencies of alpha-helix, beta-sheet and loop/turn conformations are 38 to 47 percent, 21 to 28 percent and 31 to 33 percent respectively.
Asparagine distribution in secondary structure conformations- In 3094 proteins of the human proteome, 30762 sites contained asparagine. Among these sites, 5984 sites were seen in 644 mouse proteins, 1029 sites in 103 fly proteins, 1834 sites in 179 plant proteins and 16745 sites in 756 yeast proteins. This study identified that asparagine is present in 44 to 50 percent of loop/turn region. Therefore, the asparagine percentage (40.2%) present in the alpha-helices were found to be closer to the expected percentage (42.1%). Beta-sheets were harboring lesser asparagines than alpha-helices.
NXS/T motif distribution in Secondary structures- It is observed that unannotated NXS/T motif region consisted of higher percentage of asparagine than all the asparagines present in the loop/turn conformation. The distribution of annotated NXS/T motif in all the organisms is different from that of unannotated motif. The percent asparagines of annotated motif was found higher in loops or turns.
Percentage annotated NXS/T motifs in 592 PDB human protein structures among 2284 sites
• 78% or 1779 sites - In loop/turns
• 9.7% or 222 sites - Alpha-helix
• 12.3% or 283 sites - Beta-sheet
Number of unannotated NXS/T motifs
• Human - 3779
• Mouse - 739
• Yeast - 1163
• Fly - 103
• Plant - 223
The percentage distribution of annotated NXS/T site in loops or turns is 60% while 51% of unannotated sites are seen distributed in this conformation while another related study showed the results of 75% and 71% respectively.
Analysis of N-Glycosylation sites in mouse and human proteome- The N-glycosylated NXS/T sites according to the Uniprot data are suggested as 27 percent out of all annotated NXS/T sites in human proteome. Among the membrane and secreted proteins, 53 percent of N-glycosylations is observed out of all the NXS/T sites. The present analysis indicates that out of 20,238 proteins in human proteome according to SwissProt, polymorphic sites involved in glycosylation are found to be present in 3328 proteins.
N-linked glycosylation prediction tool
The SFAT tool can carry out the tasks like
• Prediction of N-linked glycosylation regions
• Determine the conformation of secondary structure at the site of interest
• Mapping of the sequence features in UniProtKB and PDB
• Helps in the analysis of site specific quantitative structures
Prediction of N-glycosylation sites
The basic rules followed in predicting N-glycosylation sites are
- They are mitochondrial or cytoplasmic and not nuclear
- Availability of endoplasmic reticulum targeting sequence
- A site has to be exposed
- The site has to be present in the loop or turn.
There were 96 new N-glycosylation sites found in the human proteome which followed all the rules. These rules are implemented in the software based framework for enhancing the accuracy of prediction.
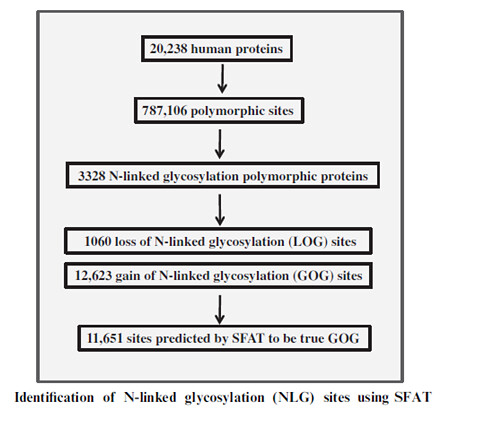
Hence, the strict rules followed potentially led to a big number of false negatives which are not as efficient as the machine based prediction. The prediction model used for cross validation showed the accuracy of NGS at 93% and the prediction of true positives was 90% of the proteins with available structures. Using the tool for the proteins without the structural information, the accuracy was evaluated as 74% and the precision was evaluated as 70%. This model was used to predict N-glycosylation sites in the polymorphic glycoproteins.
Reference:
Phuc Vinh Nguyen Lam, Radoslav Goldman, Konstantinos Karagiannis, Tejas Narsule, Vahan Simonyan, Valerii Soika, Raja Mazumder. Structure based comparative analysis and prediction of N-linked glycosylation sites in evolutionarily distant eukaryotes. Genomics Proteomics Bioinformatics 11 (2013), 96-104.
About Author / Additional Info: