The interaction between a globular protein domain and linear peptide constitutes a vital part of the cellular processes like signaling and regulatory pathways. The protein-peptide interaction has been a primary target for drug design. The functional role of a protein involves its interaction with other molecules playing an important role in various pathways. In most of the interactions involved in processes of apoptosis, immune system, and signal transduction, the proteins contain specific domains to interact and bind to the peptides. These protein-peptide interactions are observed commonly in the protein structures of proteases, major histocompatibility complex, Src Homology 2 domain, antibodies, PapD chaperone, calmodulin, and oligopeptide permeaseA. These structures were found to have different sequence specificities and binding affinities.
It is found that the interface of the protein-peptide interaction is accessible for many drug targets that are both small molecules as well as inhibiting peptides. Therefore, peptides can be designed and synthesized such that they can block the specific disease interaction. In the protein data bank or PDB, it is observed that nearly 20 new entries per month will show the interactions between protein and peptide. As the protein-peptide interaction showing structures are increasing in number, the procedures that are involved in the recognition of protein-peptide association also would improve.
Other protein-peptide Databases
In order to analyze the protein-peptide interactions, it is essential to have a database consisting of protein peptide complexes. There are certain sequence based protein-peptide interaction databases named as PhosphoELM, PepBank, ELM, DOMINO, APD, BIOPEP, SCANSITE, and ASPD. The databases like PeptiDB and PepX comprises of the structures of the protein-peptide complexes. PeptiDB constitutes a collection of 103 PDB structures that harbor non-redundant protein-peptide interactions while PepX constitutes 1431 X-ray protein structure groups centered on backbone variations and binding interfaces. There are several studies dealing with the protein domains that can interact with many other peptides.
PepBind
To study about the interactions between same proteins with different peptides a large amount of data is necessary. To resolve this issue, peptide binding protein database was created which is also called PepBind. PepBind consists of 3100 PDB protein structures without considering their method of structure determination and protein backbone similarity.
The interfaces of the protein-peptide complexes comprise of hydrogen bonds for every 100 square Angstroms of surface area that is open to the solvent. Keeping note of the relevance of the hydrogen bond, hydrophobic and ionic interactions at the protein-peptide interface, a web based interaction tool called Protein inter-chain interaction or PICI was developed. This tool will evaluate all types of interactions at the interface in the tertiary structures of the protein-peptide complexes and these interactions can be visualized further in Jmol viewer. There is another tool called “protein interaction calculator” or PIC which is designed to calculate the interactions that are explicit for the peptide chain. These can be visualized especially in the same web page accompanied by the sequence carrying the residues important in the interaction. In this database, there is a server built for binding prediction. This server helps in predicting the probable domains in the PepBind database, which can interact with the custom peptide sequence.
Results
The PepBind database aids in the exploration of the interactions between peptide and protein through computational evaluation of various interactions. It also helps in providing structural information of these interactions in the text format as well as in 3D format on the screen. PepBind database harbors the coordinates of the protein structure, protein-peptide interaction data files formed by PICI and PDBML data files. PepBind provides the structural, functional and source data for studying the interactions of peptide binding proteins. Current researchers are also allowed to submit new protein-peptide complexes which will be uploaded after appropriate authentication.
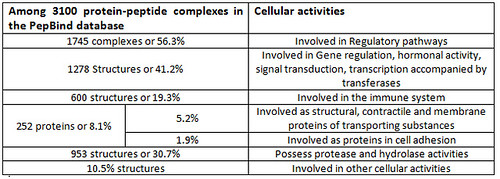
The statistical details of PepBind
Web Interface
The interface in the web regarding this database is embedded with several search tools to make the users to reach the data very easily. The “simple search” option allows the users to look for protein-peptide complexes with the help of their protein name and PDB ID. The option of “keyword search” will retrieve all the protein structures from the available fields of all the tables in PepBind bearing the query. The option of “advanced search” will help the users in retrieving the information regarding cellular function of protein, filter depending on peptide length, authors involved in protein structure determination, and structure determination methods. To search for any protein sequence homologous to the submitted sequence, PepBind can be subjected to BLAST search. The web interface reveals all the chains of the protein structure and PICI tool is used to analyze each of them. The PICI tool helps in displaying the residues involved in the interaction and are made to visualize in Jmol tool. The page that displays the protein details will have different tabs like sequence, summary, gene ontology, source, methodology, citation, external links and Ramachandran plot.
Reference:
Arindam Atanu Das, Om Prakash Sharma, Muthuvel Suresh Kumar, Ramadas Krishna, Premendu P. Mathur. PepBind: A comprehensive database and computational tool for analysis of protein-peptide interactions. Genomics Proteomics Bioinformatics 11 (2013), 241-246.
About Author / Additional Info: