Allele mining is a promising approach to dissect naturally occurring allelic variation at candidate genes controlling key agronomic traits which has potential applications in crop improvement programs.
Introduction
Identification and access to allelic variation that affects the plant phenotype is of the utmost importance for the utilization of genetic resources, such as in plant variety development. Considering the huge numbers of accessions that are held collectively by gene banks, genetic resources collections are deemed to harbour a wealth of undisclosed allelic variants. The challenge is how to unlock this variation. Allele mining is a research field aimed at identifying allelic variation of relevant traits within genetic resources collections.The process of identifying alleles of a known gene/ locus that are involved in a particular mechanism for any given trait and their variants within other genotypes or identifying novel, superior and beneficial alleles from the germplasm or natural population is known as allele mining. Allele mining is a recently developed technique that promises to detect naturally occurring allelic variation in candidate genes that regulate important agronomic trait and has potential usage in crop improvement. Allele mining allows the validation of specific gene(s) responsible for an individual trait and mining of many favorable alleles from the gene bank. DNA bank could be developed to utilize allele mining to determine unique germplasm that contains novel genes as well as allele combinations. Now different bioinformatics databases/ tools used for allele mining which is beneficial for crop improvement.
Mechanism behind formation of new alleles
An important goal of modern crop science is to use nucleotide sequence variations to improve crops. Variations can either be natural, from divergent populations, or induced through treatment with mutagens. Mutations are usually a sudden and heritable change in a gene that causes existing allelic diversity among organisms and/or any crop species. The generations of mutations in specific genes that can then be assessed for phenotypes (reverse genetics) is a powerful strategy for elucidating gene function and for creating new varieties. Mutational effects could be beneficial, dangerous, or neutral, depending on the context or even location. Mutations may take the form of insertions or deletions, which might together, called InDels, or may alter a single nucleotide inside the genome sequence, resulting in a single nucleotide polymorphism (SNP). The mutation in the coding regions could have an excellent effect on the phenotype through the changing the particular encoded healthy protein structure as well as function unlike mutations in non-coding regions of gene. Allelic variations at presented loci inside a population are usually selected and fixed within a new individual or line to produce new and important cultivars. Allele mining is expected to further increase the speed of detection of mutations and allowed rapid identification and characterization of allelic variants.
Different approaches for allele mining
There are two approaches for allele mining: (i) Eco-TILLING (Targeted Induced Local Lesions in Genomes) and (ii) Sequence based allele mining (Fig.1). Eco-TILLING is a high throughput, low cost techniques for rapid discovery of polymorphism in natural populations. Eco-TILLING is similar to targeting induced local lesion in genomes, or TILLING, but Eco-TILLING differs in that naturally occurring polymorphism are detected as opposed to experimentally induced mutations. Single nucleotide polymorphisms (SNPs), small insertions and deletions, and variations in microsatellite repeat number can be efficiently detected using the Eco-TILLING technique. Sequencing based allele mining involves PCR based amplification of alleles of a gene in varied genotypes and then DNA sequencing to recognize nucleotide variance in the alleles. Various alleles among the cultivars through this approach can be identified. The method would help to analyze individuals for haplotypes structure and diversity to infer genetic association’s studies in plants. Using this approach to recognize the effect of mutation on gene structure, the sequences are analyzed for the location of point mutation or SNPs and insertions or deletions (InDels) to construct haplotypes. Next generation sequencing (NGS) technologies can generate DNA sequence data in expensively and faster compared with traditional technologies. NGS will soon be cost effective measure for allele mining and useful for the rapid identification of important genes.
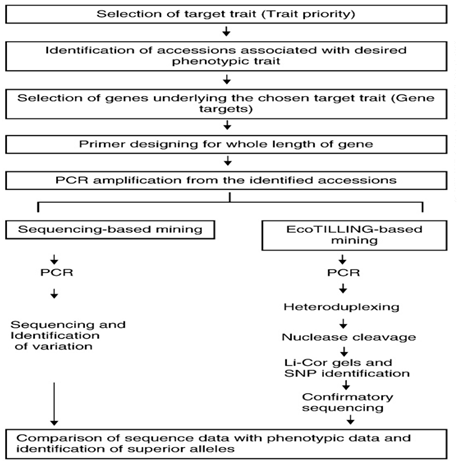
Fig 1. Different steps and Approaches in allele mining
Importance of allele mining
i. It helps in tracing the evolution of alleles
ii. Identification of new haplotypes and development of allele-specific markers for use in marker-assisted selection
iii. This capability will be important for giving breeders direct access to key alleles conferring
(1) resistance to biotic stresses,
(2) tolerance of abiotic stresses,
(3) greater nutrient use efficiency,
(4) enhanced yield, and
(5) improved quality, including human nutrition
iv. It can also provide insight into molecular basis of novel trait variations and identify the nucleotide sequence changes associated with superior alleles. In addition, the rate of evolution of alleles.
Applications of allele mining
Allele mining can be successfully employed for the detection of additional alleles from the gene of interest in diverse genetic resources. Allele mining can offer insight into the molecular basis of variations for any trait to recognize the nucleotide sequence changes related to superior alleles. Allele mining helps to distinguish molecular variation among related species. It assists in monitoring the development of alleles and identification of new haplotypes at candidate genes of interest, as well as examining the haplotype variety. Using sequence information from allele mining studies, syntenic relationships can be analyzed for the identified loci/genes over the species. Allele mining exploits the particular DNA sequence of single genotype to isolate useful alleles from related genotypes. The accessibility of sequence of the candidate genes that underlie agronomic traits may help the practical gene based use of genetic resources.
Bioinformatics Databases/ Tools used for Allele Mining
Presently, there are many bioinformatics tools and computer software programs are freely available for the identification of sequence polymorphism, comparison with other DNA sequence, prediction of amino acid changes, and identification of transcription factor binding sites (TFBSs), transcriptional regulatory elements and a database of plant promoter sequences. Some of them include PLACE, DnaSP, plantCARE, TRANSFAC, JASPAR, MEME, Plantprom DB, DCPD, and SCPD. Apart from these specialized databases/ tools there are other common bioinformatics tools which are which are helpful for gene mining. Some of them are ClustalW, MUSCLE, Probcom, T-coffee are widely used to compare and align multiple nucleotides/ amino acid sequences. FastPCR, Primer3 are tools used for In silico Primer design. MEGA package is used for phylogenetic analysis. Genebank, DDBJ, EMBL are major nucleotide databases that are used for the gene related information for different crop varieties.
References:
1. Kumar, G. R., Sakthivel, K., Sundaram, R.M., Neeraja, C.N., Balachandran, S.M., Shobha Rani, N., Viraktamath, B.C. and Madhav, M.S (2010). Allele mining in crops: Prospects and potentials. Biotechnology Advances. 28: 451â€"461
About Author / Additional Info:
I am currently pursuing Ph. D in Agricultural Biotechnology from Orissa University of Agriculture and Technology, Bhubaneswar.