Authors: Kishor U Tribhuvan1, Deepak V Pawar1, Anshul Watts1, Archana Watts2, Ravi Prakash Saini1
1ICAR-National Research Centre on Plant Biotechnology, New Delhi110012
2 ICAR-Division of Plant Physiology, Indian Agriculture Research Institute, New Delhi 110012
In the last decade utilization double stranded small RNA was started for silencing of desired genes through the phenomenon of RNA interference (RNAi). There are many types of small RNAs that have been discovered [microRNA (miRNA), Piwi-interacting RNA (piRNA), small interfering RNA (siRNA), small nucleolar RNA (snoRNAs), tRNA-derived small RNA (tsRNA), small rDNA-derived RNA (srRNA), small nuclear RNA/ U-RNA], among these miRNAs and siRNAs are rigorously utilized in RNAi gene silencing experiment. The miRNAs originate from endogenously microRNA (MIR) gene families, whereas siRNA originate exogenously in the host organism from genome of infected viruses, bacteria or other parasitic organism. With the advancement of sequencing technologies high throughput sequencing is presently being applied for sequencing of small RNA libraries and it became evident that miRNA represent only a tiny fraction of the total complement of small RNA present in the plant genome. For example in A. thaliana only ~1200 to 1300 MIR genes are present whereas more than ten thousands of small RNAs are found in inflorescence tissue. This variation between miRNA genes and number of small RNA present in the cell enforce to think about biogenesis of small RNA source.
Recent studies carried out in flies and mammals using high throughput sequencing show that endogenous biogenesis of small RNAs can take place from genes other than MIR gene family, called endogenous siRNA (endo-siRNA). These studies have also helped to understand that miRNAs are endogenously originated from MIR genes and siRNAs can be originated from pericentromeric region, transposable element and repetitive region of the genome. Transcription of MIR genes is carried out by RNA polymerase II (RNA Pol II) to produce precursor miRNA in plant and animal. In yeast (Schizosaccharomyces pombe) RNA Pol II is also carries out synthesis of siRNAs from pericentromeric repeat region and these processed siRNAs direct pericentromeric heterochromatin formation by recruiting histone methyltransferases to modify the chromatin. In case of plants, RNA polymerase IV (RNA Pol IV) and RNA polymerase V (RNA Pol V) is responsible for biosynthesis of siRNAs from pericentromeric, transposable element and other repeat regions of the genome. The subunit composition of these two polymerases indicate that these polymerases are evolved from RNA polymerase II because they share a number of subunits with RNA polymerase II. Around 90% of all endogenous siRNA are produced from RNA polymerase IV. It is thought that RNA pol IV transcribes heterochromatic DNA to produce siRNA precursor, although presence DNA dependent RNA polymerase activity is not detected so far and requires a putative chromatin remolding protein CLASSY1 for siRNA biogenesis. It is also found that majority of endogenous siRNAs is disappear in RNA pol IV mutant, the small quantity of siRNAs that exists in these mutants may be produced from other polymerase such as RNA Pol I, II and III. It is also well known that RNA Pol V is essential for siRNA mediated DNA methylation and also promotes siRNA accumulation at some loci. The main function of RNA Pol V is to methylate DNA or histone at the siRNA generating loci, promoting siRNAs biogenesis in indirect way, because DNA and histone methylation may in turn mark these regions for siRNA production in a feed forward loop. With the help of DRD1 (a chromatin remolding protein) and DMS3 (structural maintenance of chromosome hinge domain protein) RNA Pol V generates noncoding transcript which is further used for siRNA mediated heterochromatin formation. RNA Pol V is also required for siRNA mediated DNA methylation and histone H3 lysine 9 (H3K9) methylation at heterochromatic loci hence it is thought that these transcripts recruit siRNAs to the genomic loci for chromatin modification. In addition to these, RNA Pol V can also physically interact with AGO4 through its GW-WG motifs, hence RNA pol V may serve the dual function of generating transcript targeted by AGO4 bound siRNA and recruiting AGO4 and associated siRNA through physical interaction.
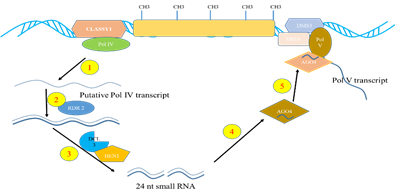
Fig. 1: Biogenesis of heterochromatic siRNAs. 1) heterochromatic loci are transcribed by RNAPol IV into ssRNA transcript, 2) ssRNA are copied into dsRNA by RDR2, 3) 24 nucleotide siRNA are processed from the dsRNA by DCL3 and methylated by HEN1, 4) Loaded in to AGO4 containing RISCs, 5) The siRNA recruited to the source loci by transcript generated by RNA Pol V
In animal system Dicer is one of the important enzyme that cleaves double stranded RNA (dsRNAs) and pre-micro RNAs into 20–25 bp dsRNA fragments and miRNAs, respectively. In plants, homolog of dicer is Dicer like (DCL) protein involved in similar kind of function. Majority of miRNAs are processed by DCL1 but in some cases it is replaced by DCL4. Whereas, DCL3 involved in processing of heterochromatic loci and produces 24 nt long siRNAs. In plant every DCL have specificity for its substrate and other DCL access the substrate in the absence of primary DCL such as production of transgene, viral and endogenous siRNA.
Argonaute (AGO) family proteins play an important role in RNAi as they are components of the RNA-induced silencing complex (RISC). These proteins bind to different classes of small non-coding RNAs, miRNAs, siRNAs and piRNAs. Small RNAs guide AGO proteins to their specific targets through sequence complementarity, which then leads to either cleavage of the mRNAs or translation inhibition. AGO proteins contain four characteristic domains: N- terminal, PAZ, Mid and a C-terminal PIWI domain. The PAZ domain is an RNA binding domain which recognizes the 3' end of both siRNA and miRNA, in a sequence independent manner and also targets the mRNA for cleavage or translation inhibition. PAZ domain bind to 2-nucleotide single stranded overhang in small RNA duplex and play a role in loading of duplex on to the protein, then the dsRNA unwound such that only one strand retained in the RISC. The PIWI domain possess RNase activity essential for small RNA guided cleavage of target mRNA. There are ten AGO protein found in A. thaliana among these AGO1 bind most of miRNAs, trans acting siRNA (ta-siRNA) and transgene siRNA and possesses slicing activity and mediate the function these small RNA in vivo. AGO1 and AGO10 are most related to each other and both are involved in miRNA mediated translational repression of target mRNA. Whereas AGO4 and AGO6 are involved for processing of endogenous siRNA to silence transposons and repeat sequences.
RNA dependent RNA polymerase (RDR) is required for amplification siRNA or miRNA, it uses ssRNAs as templates and generate ds RNA. In case of A. thaliana among the six RDR genes, RDR1, RDR2 and RDR6 have been involved in amplification of siRNA generated from plant viruses. RDR2 also a crucial player for amplification of siRNA synthesized from heterochromatic region.
References:
- Xuemei Chen (2009) Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol.35:21- 44
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ (2005) Elucidation of the small RNA component of the transcriptome. Science 309: 1567-69
About Author / Additional Info: