Author: Dr. Suresh Kaushik
Innovation in plant breeding is essential to meet the challenges of global changes such as climate change and population growth. The limited resources of land and water have to be used more efficiently for crop production as the increase in world population and the need to protect the environment. Therefore, plants with useful traits for disease, herbicide, stress tolerance, pest resistance and improved product quality characteristics have to be developed. Plant breeding started about ten thousand years ago by selecting the best plants in the field leading to domestication. The discovery of the law of genetics by Gregor Mendel about 150 years ago enhanced the speed of plant breeding. The technique of cross breeding was followed by hybrid breeding in the 1930s, cell and tissue culture methods in the 1960s and recombinant DNA techniques and genetic engineering in the 1980s. With the use of molecular markers, genome mapping and sequencing, so called smart breeding started in the late 1990s. Many genetically modified crops were developed but biotechnology companies and plant breeders are concerned about the legislative uncertainty of the genetically modified organisms (GMOs). According to the Directives 2001/18/EC, article 2(2); a GMO means an organism with the exception of human beings, in which the genetic material has been altered in a way that does not occur naturally by mating and/or natural recombination. The GMO legislation has been revised and additional legislation was introduced in 2015 to regulate food and feed derived from GMO crops. However, the definition of GMOs remains the same as in 1990. During the last 25 years new biotechnological techniques (NPBT) have been developed. They create new challenges for regulators when applying the GMO definition from 1990. Crops produced using some these NPBTs cannot be distinguished from their conventionally bred counterpart and therefore there are claims that they should be exempted from the GMOO legislation. Moreover, regulation costs for plant varieties classified as GMOs are much higher than those needed for the registration and approval of non-GM plant varieties. Many new plant breeding techniques such as cisgenesis (Fig.1 ), intragensis, near intragenesis and genome editing are currently applied as potential alternative to transgenesis to generate engineered crops with various new traits.
Cisgenesis is a genetic modification of a recipient species with a natural gene from a crossable-sexually compatible-organism (same species or closely related species). Such a gene includes its introns and is flanked by a native promoter and terminator in the normal sense orientation. Hence, in cisgenesis the T-DNA cassette is from crossable relative species or native species. The inserted gene is unchanged with its introns and regulatory sequences.
Intragenesis is the integration of an intragene which is commonly a hybrid gene. This involves the insertion of a reorganized, full or partial coding part of a natural gene frequently combined with another promoter and/or terminator from a gene of the same species or a crossable species. Thus, the inserted DNA can be a new combination of DNA fragments from the species itself or from a cross-compatible species and the transferred T-DNA cassette including the coding sequence and regulatory elements is taken from other cross-incompatible species. While the DNA cassette in intragenesis is in vitro rearrangement with genetic elements from both host and cross-compatible donors but in near intragenesis the T-DNA cassette consists of host derived genetic elements with minimal recombinant elements (Singh et al. 2015). Cisgenic and intragenic plants are produced by the same transformation techniques as transgenic plants e.g. Agrobacterium-mediated transformation, following the isolation of genes from the host. Biolistic method could be used for the transformation. With Agrobactrium-mediated transformation the vectors used may contain Agrobacterium T-DNA border sequences to facilitate the insertions of the target genes into the plant genome. Therefore, the resulting plants might contain small, non-coding bacterial sequences. However, specific vector have been constructed for cisgenic/intragenic approaches which use DNA sequences originating from the same crop species or related species to inset the target genes. Intragenesis and cisgenesis use the same gene pools available for traditional breeding and no foreign genetic elements are present in the end products, but potentially interrupting and silencing the resident genes or other host sequences due to random integration can take place. Irrespective of whether the cisgenic or intragenic approach is used there exists a possibility that the insert interrupt open reading frame (ORF) in the host plant or create new ones as a consequence of the insertion process. Deletions of host DNA can also occur following insertion.
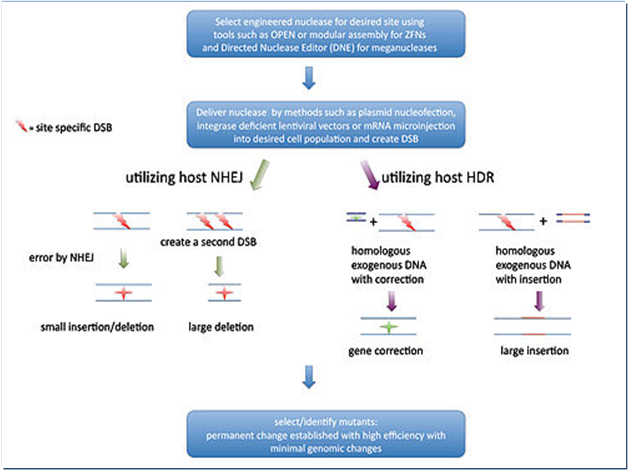
Figure I. Outlines of conventional, transgenesis and cisgenesis breeding techniques (Source: Wikipedia)
Genome editing is a highly specific and efficient tool with a great potential to generate new improved crops. Genome editing is based on site-specific mutagenesis mediated by engineered nucleases. The most commonly used engineered nuclease (Fig. 2) are zinc finger nucleases (ZFN), meganucleases, transcription activator-like effectors nuclease (TALEN) and clustered regular interspaced short palindromic repeats/CRISPR-associated nuclease (CRISPR/Cas) (Araki et al. 2014; Cardi and Varshney, 2016; Jones, 2015).
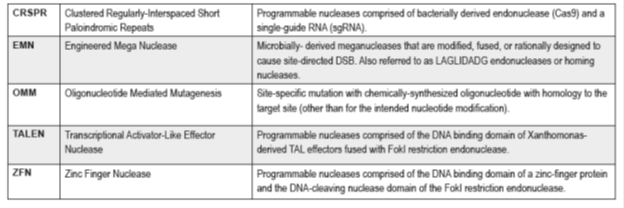
Figure 2. Current genome editing technology used for generation of improved crops (Source: Wolt JD et al., 2015)
An engineered nuclease initially create a double strand break (DSB) in the target sequence and the DSB can be repaired inaccurately in vitro through two mechanisms (Fig.3): firstly, an error prone process named non-homologous end joining (NHEJ) that often generate indels (insertions or deletions) and secondly, a homologous DNA recombination (HDR) process that results in replacement/insertions or deletion depends on the presence or absence of a donor DNA, respectively (Pauwels et al., 2014).
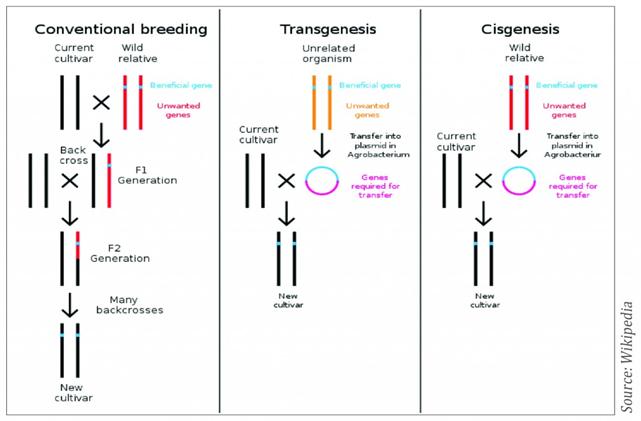
Figure 3. Mechanisms of Genome Editing
Source: https://en.wikipedia.org/wiki/Genome_editing
An engineered nuclease gene is stably integrated into the host genome initially but it is lost in the next generation due to random assortment and chromosome segregation (Jone, 2015). Hence crops generated by genome editing are virtually indistinguishable from those generated by natural mutation or conventional mutagenesis. Genome editing produces crops with indels in a targeted gene involving changes in deletion or insertions of a few nucleotide bases only and making it difficult to distinguish them from natural variation. Insert DNA fragments via homologous recombination into the host genome at pre-determined locations, off-mutagenesis and indented monoallelic modification could still occur (Araki et al., 2014; Jones, 2015; Pauwels et al., 2015; Wolt et al., 2016).
Targeted NPBT minimizes the probability of position effects and unintended gene disruption which is common in transgenesis due to random insertion of foreign genetic materials. Hence, risk induced by NPBT cannot be ruled out. The application of these new techniques have raised considerable concern debate on their regulation (Lusser and Davies, 2013; Broeders et al., 2013). Current regulation on crops safety put great emphasis on the regulation of genetically modified crops because of concerns such as co-introduction of selectable markers particularly antibiotic resistance genes and microorganism originated DNA sequences or unintended effects caused by random insertion, but in case of genome editing which acts on the whole gene sequence, including promoter sequence, off-target genome editing could occur. This would affect the host both genetically and epigenetically. Therefore, crops produced by this method still need proper screening and characterization. Since, more and more engineered crops derived from NPBT are in the pipeline to be commercialized and released to environment in the near future (James, 2016); it creases great challenges for molecular characterization to provide risk assessment study.
Conclusions
NBPT offers new opportunity to transfer genetic materials from closely related species or native genetic pools into a host by precise site-directed mutagenesis. This eliminates the introduction of foreign genetic elements. NPBT may accelerate plant breeding process because it relieves to some extent the worries from regulators and public about the undesirable effects due to random gene insertions and disruptions associated with conventional GM crops. Still, we cannot rule out the effects caused by random occurrence of unintended outcome associated with NPBT. Hence, sufficient risk assessment of crops generated by NPBT is still essential at DNA, RNA, protein and metabolic levels using genomic, transcriptomic, proteomic and metabolic study, respectively.
References:
- Araki M et al., 2014. Caution required for handling genome editing technology. Trends Biotech. 32: 234-237
- Broeders SR et al., 2012. How to deal with the upcoming challenges in GMO detection in food and feed. J. Biomed. Biotech. 2014, 402418.
- Cardi T, Varshney R. 2016. Cisgenesis and genome editing: combining concepts and efforts for a smarter use of genetic resources in crop breeding. Plant Breed. 135, 139-147.
- Directive 2015/412, new rules formally approved on 2 March 2015, http://www.consilium.europa.eu/en/press/press-releases/2015/03/150302-new-gmo-rules-get-approved.
- James C, 2016. 20th Anniversary of the Global Commercialization of Biotech Crops (1996 to 2015) and Biotech Crop Highlights in 2015. ASAAA Briefs 51.
- Jones HD 2015. Regulatory uncertainty over genome editing. Nat. Plants 1, 14011.
- Lusser M, Davies HV 2013. Comparative regulatory approaches for groups of new plant breeding techniques. Nat. Biotech. 30: 437-446.
- Pauwels K et al. 2014. Engineering nucleases for gene targeting: safety and regulatory considerations. Nat. Biotechnol. 31: 17-27.
- Singh A et al. 2015. Alternative to transgenesis: cisgenesis and intragenesis. Advances in Plant Breeding Strategies: Breeding Biotechnology and Molecular Tools. Springer International Publishing. Pp. 345-367.
- Wolt JD et al. 2016. The regulatory status of genome-edited crops, Plant Biotechnol. J. 14, 510-518.
About Author / Additional Info:
Working as Chief Technical Officer (Research) at Indian Agricultural Research Institute, Pusa, New Delhi with Ph.D. in Molecular Biology and Biotechnology from IAIR, New Delhi, India.
http://in.linkedin.com/in/sckaushik