Author: Helan Baby Thomas
Department of Plant Biotechnology, Indira Gandhi Agricultural University, Raipur-492012
Correspondance Address: hthomas392@gmail.com
Introduction
Forward genetics refers to the process of determining the genetic underpinnings of observable phenotypic variation. In many cases the observable variation has been induced using a DNA damaging agent (mutagen) but also may be naturally occurring. The investigator eventually ends up sequencing the gene or genes thought to be involved.
The advancement of whole genome sequencing has enabled many researchers to have access to all of the gene sequences within a given organism and their function. So, instead of going from phenotype to sequence as in forward genetics, reverse genetics works in the opposite direction – a gene sequence is known, but its exact function is uncertain. In reverse genetics, a specific gene or gene product is disrupted or modified and then the phenotype is measured. Some of the reverse genetics techniques are explained below:
Virus Induced Gene Silencing (VIGS)
Virus-induced gene silencing (VIGS) has emerged as one of the most adaptable techniques in recent years but its effects are transient in nature. VIGS has been used widely in plants for analysis of gene function and has been adapted for high-throughput functional genomics. VIGS is a technology that exploits an RNA-mediated antiviral defense mechanism (Lu et al., 2003). The process is initiated by cloning a 200–1300 bp cDNA fragment from a plant gene of interest into a DNA copy of the genome of an RNA-virus and transfecting the plant with this construct using Agrobacterium. Double-stranded RNA from the viral genome, including the sequence from the gene of interest, is formed during viral replication. The double-stranded RNA molecules are degraded into small interfering RNA (siRNA) molecules by the plant Dicer-like enzymes, thus activating the siRNA silencing pathway. This results in the degradation of the target gene transcript leading to a knockout or knockdown phenotype for the gene of interest.
Insertional mutagenesis
This technology has been used in both monocotyledonous and dicotyledonous plants for insertional knockouts. A population of plants each having an insertion at a unique site in the
genome is generated either by transformation (T-DNA) or transposon activation. Screening the population for finding the insertion in the gene of interest can be done with PCR using one gene-specific primer and one insertion based primer. Gene disruption studies using this technique typically result in a total loss of function and the insertion can be easily followed, using PCR, in a population of plants where it is segregating.
RNA mediated Interference (RNAi)
RNAi is a powerful, conserved biological process through which the small, double-stranded RNAs specifically silence the expression of homologous genes, largely through degradation of their cognate mRNA. In this procedure, a microRNA (miRNA) gene carrying a 21 bp insert complementary to the target gene is transformed into the plant where it is processed by the Dicer pathway to produce the small silencing RNA molecules that target endogenous transcript(s) for degradation. The process of RNAi is described in Fig 1.1.
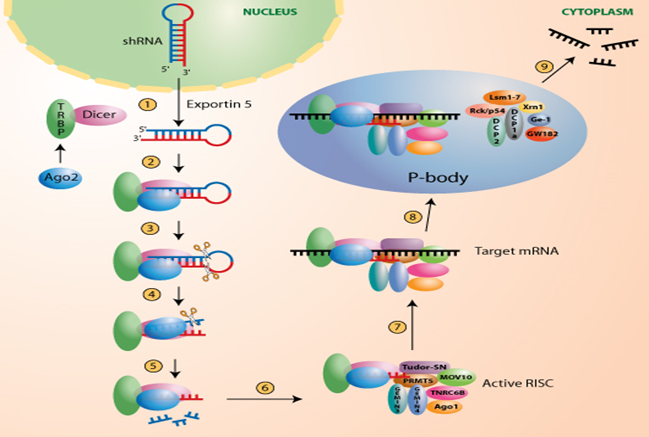
FIG.1.1. Process of RNAi (Lu 2005, Jakymiw 2005).
TILLING
TILLING (Targeting Induced Local Lesions IN Genomes) is a non-transgenic reverse genetic technique that is suitable for most plants . Steps in TILLING are given (Fig.1.2):
- Developing a mutagenized population
- Sample DNA pooling
- Mutation Discovery
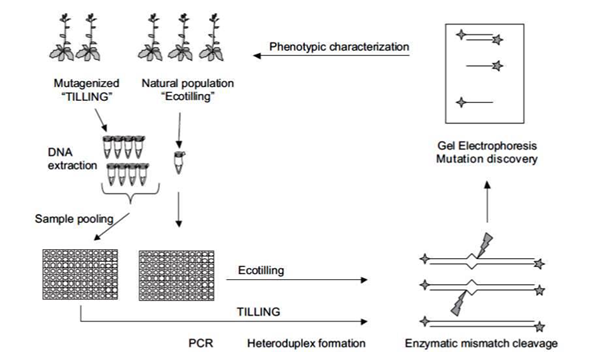
Technique of TILLING (Bradley et al., 2007)
Conclusion
Reverse is one of the most efficient tools to identify the genes and their function. With the continuous development of new technologies, the most cost-effective technique for examining gene function in plants in the future may involve direct sequencing of part or complete genomes or some other genome analysis technique that is yet to be developed.
References
Bradley J. Till, Luca Comai and Steven Henikoff (2007). Tilling And Ecotilling For Crop Improvement. Genomics-Assisted Crop Improvement: Vol. 1: Genomics Approaches and Platforms, 333–349.
Burch-Smith TM, Anderson JC, Martin GB, et al. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 2004; 39:734–46.
Jakymiw A. et al., 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 7(12):1167-74.
Liu J. et al., 2005. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 7(12):1161-6.
Rui Lu, Ana Montserrat Martin-Hernandez, Jack R. Peart, Isabelle Malcuit, and David C. Baulcombe (2003). Virus-induced gene silencing in plants. Science Direct Methods 30 (2003) 296–303
Stepanova AN, Alonso JM. PCR-Based Screening for Insertional Mutants. Methods Mol Biol 2006; 323:163–72.
About Author / Additional Info:
I am currently pursuing PhD in Biotechnology from Indira Gandhi Agricultural University, Raipur