Authors: Rohini Bansode1 and Sandeep Kumar2
1 Department of Plant Biotechnology, KAU, Kerala-680656
2 Indian Agricultural Research Institute, New Delhi 110012
Nature uses both mutation and recombination to increase the number of combinations of gene. To recreate evolution in the laboratory, the mechanisms of natural evolution must be accelerated such that meaningful diversity can be created and selected in a much shorter timeframe, mere days to weeks being favored. This defines the two-fold strategy of directed evolution:
a) Rapid generation of a functionally diverse collection of mutants.
b) Rapid identification of the best performers among them.
1. DNA Shuffling
DNA shuffling refers to in vitro homologous recombination of pools of selected genes by random fragmentation and polymerase chain reassembly. This method is introduced by Stemmer in 1994. It involves the digestion of a gene by DNaseI into random fragments, and the reassembly of those fragments into a full-length gene by primerless PCR: the fragments prime on each other based on sequence homology, and recombination occurs when fragments from one copy of a gene anneal to fragments from another copy, causing a template switch, or crossover event.
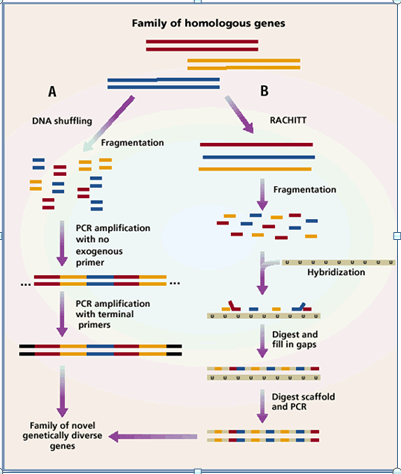
Image Source: http://www.lookfordiagnosis.com/mesh_info.php?term=Dna+Shuffling&lang=5 | nature.com
Advantages of DNA shuffling
• Error-prone PCR can be used to mutagenize a mixture of fragments of unknown sequence. The published error-prone PCR protocols do not allow amplification of DNA fragments >0.5-1.0 kb, limiting their practical application.
• This method can be applied to sequences >1 kb.
• It has a mutagenesis rate similar to error-prone PCR.
• It also works with pools of unknown sequence.
• This method makes possible to remove neutral mutations by backcrossing with excess parental or wild-type DNA.
2. Family Shuffling Method
It refers to the recombination of a family of related genes from various species. This method is introduced by Crameri et al., 1998. It involves mixing of a family of homologous sequences obtained from nature, typically the same gene from related species or related genes from a single species.
3. Random Chimeragenesis on Transient Templates (RACHITT)
This method does not utilize thermocycling, strand switching, or staggered extension of primers. Instead it uses trimming, gap filling and ligation of parental gene fragment hybridized on a transient DNA template. Instead, a uracil-containing parent gene is made single-stranded to serve as a scaffold for the ordering of top-strand fragments of additional, homologous parent gene(s), and recombination occurs when fragments from different parent genes hybridize to the scaffold. Pfu DNA polymerase 3'-5' exonuclease activity removes the unhybridized 5' or 3' overhanging "flaps" created by fragment annealing, and also fills gaps between the annealed fragments using the transient scaffold as a template. The template strand is then eliminated by treatment with uracil-DNA-glycosylase before applying the template-chimera hybrid to PCR, resulting in amplification of double stranded, homoduplex chimerical gene sequences.
Advantages:
1. It provides a significantly higher rate of crossover compared to other family shuffling methods, with an average of 14 crossovers per gene versus one to four crossovers for most other methods.
2. It also generates 100% chimerical progeny with no duplications of recombination pattern in chimerical genes.
3. This method utilizes single hybridization step because it lends to greater binding specificity for DNA fragment of both low and high sequence identity and resulted in no unshuffled parental clones.
Applications of DNA shuffling
1. Applications in enzyme engineering
a) Improving catalytic activity eg. Subtilisins, Thermostable p-Nitrobenzyl esterase
b) Expanding specificity eg. E. coli D-2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase
2. Applications in pathway engineering eg. Carotenoids
3. Applications in protein engineering eg. Polyketide synthesis, Green fluorescent protein, Fucosidase, Thymidine kinase, Beta-lactamase etc.
4. Agriculturally important traits eg. Glyphosate resistance. Bacillus thuringiensis toxin.
References:
1. Crameri A., Raillard S.A., Bermudez E and Stemmer W.P., 1998, DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nat Biotechnol, 319: 536-538.
About Author / Additional Info:
I am currently pursuing Ph.D in Plant Biotechnology from Kerala Agricultural University, Thrissur.