Authors: Rajani Verma1 and M.L. Jakhar2
1Department of Plant Breeding and Genetics, SKNAU, Jobner 303329 (Raj.) (India)
2Professor & Head Department of Plant Breeding and Genetics, SKNAU, Jobner 303329 (Raj.) (India)
What is DNA Sequencing
DNA Sequencing refers to the process of recording the exact sequence of nucleotides in a DNA segment of an organism corresponding to a specific gene. Gene Sequencing is a process of determine the nucleotide order of a given DNA fragment, called DNA Sequencing. DNA sequencing is the process of determining the sequence of nucleotide bases (As, Ts, Cs, and Gs) in a piece of DNA.
Main points related to DNA Sequencing:
- The DNA sequencing is a useful in both basic and applied research in biological science especially in molecular biology.
- The gene Sequencing can be done for a specific gene as well as for the entire genome.
- The knowledge of DNA Sequencing is very much useful in medical science. It can be used for identification, diagnosis and treatment of genetic disease.
- In plants, knowledge of gene sequencing will be help in controlling disease and improving quality of the product and resistance to biotic and abiotic stresses.
- DNA Sequencing also referred to as Gene Sequencing or nucleotide sequencing.
Sanger sequencing: The chain termination method
This method was developed by Frederick Sanger in 1977. Sanger sequencing method also known as chain termination method. In this method a low concentration of a chain terminating nucleotide, commonly known as dideoxy nucleotide is used. Hence this method is also referred to as dideoxy DNA Sequencing procedure.
Ingredients for Sanger Sequencing :
- The template DNA to be sequenced
- A DNA polymerase enzyme
- A primer, which is a short piece of single-stranded DNA that binds to the template DNA and acts as a "starter" for the polymerase
- The four DNA nucleotides (dATP, dTTP, dCTP, dGTP)
- Dideoxy, or chain-terminating, versions of all four nucleotides (ddATP, ddTTP, ddCTP, ddGTP), each labeled with a different color of dye
Method of Sanger sequencing The DNA sample to be sequenced is combined in a tube with primer, DNA polymerase, and DNA nucleotides (dATP, dTTP, dGTP, and dCTP). The four dye-labeled, chain-terminating dideoxy nucleotides are added as well, but in much smaller amounts than the ordinary nucleotides.
The mixture is first heated to denature the template DNA (separate the strands), then cooled so that the primer can bind to the single-stranded template. Once the primer has bound, the temperature is raised again, allowing DNA polymerase to synthesize new DNA starting from the primer. DNA polymerase will continue adding nucleotides to the chain until it happens to add a dideoxy nucleotide instead of a normal one. At that point, no further nucleotides can be added, so the strand will end with the dideoxy nucleotide.
This process is repeated in a number of cycles. By the time the cycling is complete, it’s virtually guaranteed that a dideoxy nucleotide will have been incorporated at every single position of the target DNA in at least one reaction. That is, the tube will contain fragments of different lengths, ending at each of the nucleotide positions in the original DNA (see figure below). The ends of the fragments will be labeled with dyes that indicate their final nucleotide.
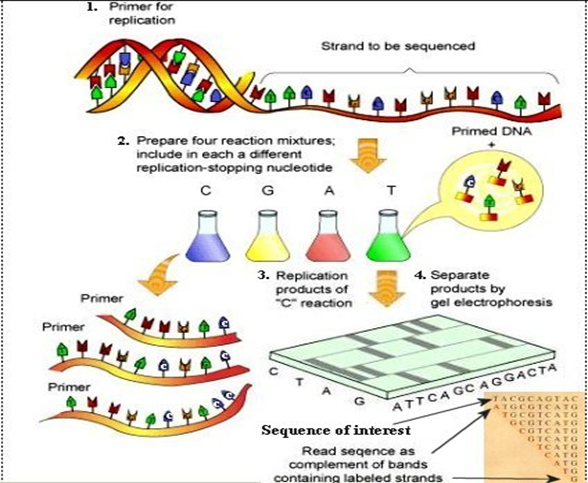
(Image Source: https://universe-review.ca/R11-16-DNAsequencing.htm)
After the reaction is done, the fragments are run through a long, thin tube containing a gel matrix in a process called capillary gel electrophoresis. Short fragments move quickly through the pores of the gel, while long fragments move more slowly. As each fragment crosses the “finish line” at the end of the tube, it’s illuminated by a laser, allowing the attached dye to be detected. The smallest fragment (ending just one nucleotide after the primer) crosses the finish line first, followed by the next-smallest fragment (ending two nucleotides after the primer), and so forth. Thus, from the colors of dyes registered one after another on the detector, the sequence of the original piece of DNA can be built up one nucleotide at a time. The data recorded by the detector consist of a series of peaks in fluorescence intensity. The DNA sequence is read from the peaks in the chroatogram.
Limitation of Sanger Sequencing Method
- The primer used can also be annealed to a second site. This will cause two sequences for interpretation at the same time.
- Sometime RNA contaminates the reaction, which can act like a primer and lead to bands in all lanes at all possible due to non-specific priming.
- Secondary structure of DNA being read by DNA polymerase can lead to reading problem.
- It can sequence only short length of DNA at a time. It can sequence maximum of about 1000 base pairs.
About Author / Additional Info:
I am currently pursuing Ph.D in PLANT BREEDING AND GENETICS from University of SKNAU, JOBNER, JAIPUR.