Author: S.T. Waghmare*, N. S. Chavan., S.A. Belge., P.T. Yeole
Sandeshwaghmare174@gmail.com
1. Introduction
Plants represent a rich source of nutrients for many organisms including bacteria, fungi, protists, insects, and vertebrates. Although lacking an immune system comparable to animals, plants have developed a stunning array of structural, chemical, and protein-based defenses designed to detect invading organisms and stop them before they are able to cause extensive damage. The plants defence mechanism are of two types i.e.,
1) Innate immunity - Cells and mechanisms that defend the host from infection by other organisms in a non specific manner
2) Adaptive immunity - Cells recognize and respond to invading pathogens in a specific manner
1.1 TAL (Transcription Activator Like) effectors
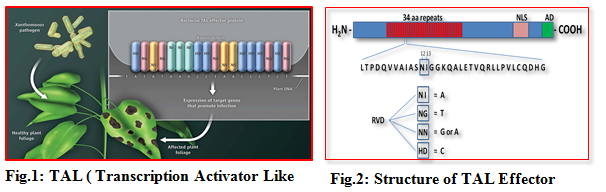
Plant immune responses rely on the recognition of conserved microbial molecules, including bacterial flagellin and elongation factor (EF-Tu), called PAMPs/MAMPs (pathogen/microbe-associated molecular patterns) by plant extracellular PRRs (pattern recognition receptors). This recognition, historically known as basal defense, results in PTI (PAMP-triggered immunity).
Transcription Activator Like Effectors (TALEs) represent a group of effector proteins that are of bacterial origin and are translocated into host cells via a secretion system during pathogen infection. TALE contains a central repetitive region consisting of a variable number of 34 amino acid repeats that are nearly identical except for the two variable amino acids at the positions 1 2 and13 known as Repeat Variable Diresidues (RVDs). A combination of repeat number and composition of RVDs determine the specificity of each TALE for its target DNA sequence. The recognition of DNA target by the TALE repeat domains appear to follow a simple code, i.e., one repeat corresponds to one nucleotide and one type of RVDs preferentially recognizes one type of four nucleotides of target.
1.2 Role of TAL effectors in pathogenesis
TALEs can be detected in the host cytoplasm as exemplified by the tomato NB-LRR type resistance (R) protein Bs4, which is capable of detecting the full length TALE AvrBs4 as well as deletion derivatives that lack nuclear localization signals. Upon interaction with importin a TALEs translocate to the nucleus, bind to matching UPT boxes, and transcriptionally activate matching host susceptibility (S) genes such as UPA20, Os8N3, OsTFIIAg1, or OsTFX1, resulting in disease. Activation of S genes can be suppressed either by a variation in the RNA II polymerase complex such as xa5 or by mutations in the individual UPT boxes (xa13 mutation).
2. Mechanisms of plant evolved disease resistance to TAL effectors
Recognition of pathogen derived molecules is a common mechanism of plant immunity and can occur outside or inside plant cells. TAL effectors are known to be translocated by the type III secretion system into the cytoplasm of host plant cells. Research has revealed that at least three different strategies exist by which resistant plants sense the presence of TAL effectors and react to them.
2.1 Recognition of TAL Effector Structure by R Genes
Recognition of pathogen effectors commonly occurs by plant disease resistance proteins harboring nucleotide binding site - leucine rich repeat (NBS-LRR) domain structures; however, TAL effectors are rarely recognized by this mechanism. To date, only one NBS-LRR has been reported to recognize the presence of TAL effectors, the tomato Bs4 protein. Bs4 has an N-terminal TIR (Toll-interleukin 1 receptor) domain and confers the ability to recognize multiple TAL effectors, such as AvrBs4, Hax3, and Hax4. Bs4 seems to be prevalent in cultivated tomatoes, and its isolation required introgression of a non functional allele that harbors a splice site mutation from Solanum pennellii LA2963, a wild relative of tomato.
2.2 Suppression of virulence functions
Another strategy for plant immunity is to abolish TAL effector function inside the plant. TAL effectors require nuclear import, DNA binding, and subsequent transcriptional activation of plant genes to exert their pathogenic potential. They are dependent on conserved plant helper components and mechanisms, such as importin-mediated nuclear localization. Infrequently, plants have variants of these helper components that are insensitive to the action of TAL effectors. For example, rice with a recessive xa5 allele of the gamma subunit of transcription initiation factor IIA (TFIIAγ), a key component of the transcription preinitiation complex, is resistant to X. oryzae strains carrying the TAL effector gene Avrxa5.
2.3 Promoter - traps and executor genes
The most common strategy for TAL effector recognition in resistant host genotypes makes use of trap promoters coupled to an executor-type resistance gene (E gene) (Plate 5 c). Induction of executor genes triggers a localized cell death response known as the HR, a common and effective mechanism of plant immunity against various pathogens. Trap promoters have evolved recognition sites for specific TAL effectors so that, upon infection, the introduction of that TAL effector trips expression of the executor gene and limits pathogen growth.
3. Mechanisms of engineered disease resistance based on TAL effectors
Using the knowledge of TAL effector biology and the naturally evolved resistance mechanisms in plants described in the previous section, scientists have begun to design and test novel mechanisms of resistance. Moreover, these approaches further exploit the unique nature of TAL effectors by making use of the toolbox of genome and transcriptome modification tools exemplified in. Below is a discussion of ways in which TAL effectors might be used to engineer plant disease resistance by exploiting mechanisms of TAL effector recognition of host genes and host recognition of TAL effectors, and by using TAL effector DNA binding domains fused to other functional protein domains such as nucleases.
3.1 Recognition of TAL effector by mutant R genes
The NBS-LRR disease resistance protein Bs4 mediates recognition of multiple unrelated TAL effectors. Strikingly, a truncated AvrBs4 variant lacking NLSs and most of the repeat region also triggers an HR in a Bs4 genetic background . This suggests that physical features rather than transcription activation trigger Bs4 perception of TAL effectors. The Bs4 protein is also capable of recognizing AvrBs3 if present at high enough levels when expressed under the strong constitutive 35S promoter.
3.2 Suppression of virulence by mutate promoters
By identifying particular TAL effectors required for virulence, strategies can be employed to impair their manipulation of susceptibility genes. Historically, evidence for genes targeted by TAL effectors has come from identification of recessive resistance genes after screening the natural diversity of a plant species. Screening for recessive resistance depends on the availability of natural variants in a collection of germplasm. Integration of computational and transcriptional approaches for the identification of TAL effector targets allows the researcher to quickly focus on candidate EBE boxes in the promoters of virulence targets.
3.3 Elaborated promoter traps
An understanding of the mechanism of the promoter-trap executor gene resistance of Xa27 and Bs3 lends itself to a simple extension of this principle: to engineer resistance by introducing one or more synthetic EBEs into an executor gene promoter to trigger resistance against pathogens that inject TALEs into plants cell during infection.
3.4 New executor genes
Compared with NBS-LRR resistance genes, which can number into the thousands in some species, far fewer examples are known of disease resistance conferred by effector-triggered transcriptional induction of executor genes. However, a recent report should pave the way for more executor-type genes to be discovered. The Bs4C gene from Capsicum pubescens was identified and cloned using RNA-Seq by capitalizing on the unique expression pattern of the gene (note that this gene and its resistance mechanism are distinct from Bs4 from tomato, the NBS-LRR gene discussed above).
3.5 Novel resistance mechanism
3.5.1 Genome modification
TALE nucleases (TALENs), fusion proteins of the full-length or truncated TALEs and the DNA cleavage domain of FokI endonuclease, are newly developed and highly efficient tools for targeted gene modification. Like zinc finger nucleases, fusion proteins of zinc finger motifs and the DNA cleavage domain of FokI, TALENs also work as the dimer by binding to two adjacent effector binding elements (EBEs), which are separated by a spacer.
5. Applications of TAL effectors
1. TAL effectors fusions is used for genome editing
2. TAL effectors based control implemented for gene expression
3. TAL effectors used for Metabolic engineering
4. TAL effectors used in study of System biology
5. TAL effectors used in Crop and livestock improvement
References:
1. Bogdanove, J., Doyle, E. L., Stoddard, B. L., Voytas, D. F., and Adam, J. 2013. TAL effectors: highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 23: 390-398.
2. Bogdanove, A. J. and Voytas, D. F. 2011. TAL effectors: customizable proteins for DNA targeting. Science 333: 1843-1846.
3. Morbitzer, R., Romer, P., Boch, J., and Lahaye, T. 2010. Regulation of selected genome lociusing denovo engineered transcription activator-like effector (TALE) Nat. Acad. Sci. USA 107: 2160-2162.
4. Schornack, S., Moscou,M. J., Ward, E. R., and Horvath, D. M. 2013. TAL effectors: finding plant genes for disease and defense. Annu. Rev. Phytopathol. 51: 383-406.
About Author / Additional Info:
I am currently working as Assistant professor at K.K. Wagh College of Agricultural Biotechnology, Nashik