Authors: *Sohan lal kajla, Mukesh Bhakal and Neelu kumari
*Department of Plant Breeding & Genetics,
S.K.R.A.U
Bikaner-334006, Bikaner, Rajasthan, India
*Email: kajlasikar@gmail.com
RNAi
- RNA interference (RNAi) is an evolutionally highly conserved process of post-transcriptional gene silencing (PTGS) by which double stranded RNA (dsRNA) causes sequence-specific degradation of mRNA sequences.
- It was first discovered in 1998 by Andrew Fire and Craig Mello in the nematode worm Caenorhabditis elegans and later found in a wide variety of organisms, including mammals.
miRNA and siRNA
| miRNA | siRNA |
| MicroRNAs(miRNAs)are genomically encoded non-coding RNAs that help regulate gene expression, particularly during development. | The siRNAs (small interfering RNA) derived from long dsRNA precursors. |
| miRNAs must first undergo extensive post-transcriptional modification | siRNAs typically base-pair perfectly and induce mRNA cleavage only in a single, specific target |
| Typically have incomplete base pairing to a target and inhibit the translation of many different mRNAs with similar sequences. | ------- |
Features of RNAi
- dsRNA needs to be directed against an exon, not an intron in order to be effective
- homology of the dsRNA and the target gene/mRNA is Required
- targeted mRNA is lost (degraded) after RNAi
- the effect is non-stoichiometric; small amounts of dsRNA can wipe out an excess of mRNA (pointing to an enzymatic mechanism)
Interesting aspects of RNAi
- dsRNA, rather than single-stranded antisense RNA, is the interfering agent.
- It is highly specific
- It is remarkably potent (only a few dsRNA molecules per cell are required for effective interference).
- The interfering activity (and presumably the dsRNA) can cause interference in cells and tissues far distant from the site of introduction.
Mechanism of RNA interference
- First it was observed by Craig Mello and Fire et al. reported in ('98), that only dsRNA targeting exon sequences was effective (promoter and intron sequences could not produce an RNAi effect).
- Additional evidence supporting mature messages as the most likely target of RNA-mediated interference is summarised below (from Montgomery et al. (98) PNAS 95: 15502-07):
- primary DNA sequence of target appears unaltered
- initiation and elongation of transcription appear unaffected
This observation suggested that the dsRNA must be either replicated and/or function catalytically.
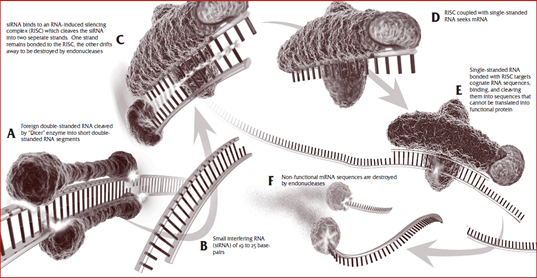
- Genetic and biochemical studies involving plants and flies as well as worms have uncovered similar processes in which the dsRNA is cleaved into ~23 bp short interfering RNAs (siRNAs) by an enzyme called Dicer.
- Thus producing multiple “trigger” molecules from the original single dsRNA.
- The siRNA-Dicer complex recruits additional components to form an RNA-induced Silencing Complex (RISC) in which the unwound siRNA base pairs with complementary mRNA, thus guiding the RNAi machinery to the target mRNA resulting in the effective cleavage and subsequent degradation of the mRNA.
- In this way, the activated RISC could potentially target multiple mRNAs, and thus function catalytically.
Applications of RNAi
1. Gene Knockdown
- Double-stranded RNA is synthesized with a sequence complementary to a gene of interest and introduced into a cell or organism, where it is recognized as exogenous genetic material and activates the RNAi pathway.
- Using this mechanism, researchers can cause a drastic decrease in the expression of a targeted gene. Studying the effects of this decrease can show the physiological role of the gene product.
- Since RNAi may not totally abolish expression of the gene, this technique is sometimes referred as a "knockdown",
Mouse oocytes and cells from early mouse embryos lack this reaction to exogenous dsRNA and are therefore a common model system for studying gene-knockdown effects in mammals and Arabidopsis in case of plants.
2. Functional genomics
- Most functional genomics applications of RNAi in animals have used C. elegans and Drosophila, as these are the common model organisms in which RNAi is most effective.
- Functional genomics using RNAi is a particularly attractive technique for genomic mapping and annotation in plants because many plants arepolyploid, which presents substantial challenges for more traditional genetic engineering methods.
RNAi has been successfully used for functional genomics studies in bread wheat (which is hexaploid) as well as more common plant model systems Arabidopsis and maize.
3.Medicines
- It may be possible to exploit RNA interference in therapy.
- Among the first applications to reach clinical trials were in the trea tment of mascular degeneration and respiratory syncytial virus.
- knockdown of host receptors and coreceptors for HIV
- RNA interference is also often seen as a promising way to treat cancer by silencing genes differentially upregulated in tumor cells or genes involved in cell division.
4.Biotechnology
- RNA interference has been used for applications in biotechnology, particularly in the engineering of food plants that produce lower levels of natural plant toxins.
- For example, cotton seeds are rich in dietary protein but naturally contain the toxic terpenoid product gossypol, making them unsuitable for human consumption.
- RNAi has been used to produce cotton stocks whose seeds contain reduced levels of delta-cadinene synthase, a key enzyme in gossypol production, without affecting the enzyme's production in other parts of plant.
- Similar efforts have been directed toward the reduction of the cyanogenic natural product linamarin in cassava plants
5.RNAi gene therapy
- Viral infections
- Human infection with viral pathogens presents an excellent target for therapeutic RNAi for several reasons.
An important example comprises viral hepatitis infections for which the pathogenic agents (e.g., hepatitis A, B, C, or delta virus) are genetically diverse.
- Cancer
- A second promising target for RNAi gene therapies is human cancer.
In general, there are three classes of RNAi targets for cancer therapies: genes that are part of cancer-associated cellular pathways, those that play a role in tumor-host interactions, or those that mediate resistance to chemo- or radiotherapy.
6. Genome-scale RNAi screening
- The genome-scale RNAi research relies on the high-throughput screening (HTS) technology. The RNAi HTS technology allows genome-wide loss-of-function screening and is broadly used in the identification of genes associated with specific biological phenotypes.
- One of the major advantages of the genome-scale RNAi screening is its ability to simultaneously interrogate thousands of genes
- The basic process of cell-based RNAi screening includes (i) the choice of an RNAi library, (ii) the selection of a robust and stable type of cells, (iii) the transfection of the selected cells with RNAi agents from the chosen RNAi library, (iv) necessary treatment or incubation, (v) signal detection, (vi) statistical and bioinformatics analysis, and (vii) the determination of important genes or therapeutical targets .
References:
- * Napoli (1990), Guo and Kempheus (1995), Cogoni (1999) and Rondinone (2006 )
- Divya Khanna et al.,2007
About Author / Additional Info:
I am currently pursuing Ph.D. in Agriculture(Department of Plant Breeding & Genetics) from SKRAU, Bikaner.
Ph.D. in Agriculture(Department of Plant Breeding & Genetics) from SKNAU(Jobner)