Author: Rahul M. Phuke
Nested association mapping (NAM) as a genome wide complex trait dissection strategy that integrates the advantages of linkage analysis and association mapping in a single, unified mapping population.
It is first designed by the labs of Edward Buckler, James Holland, and Michael McMullen for identifying and dissecting the genetic architecture of complex traits in corn (Zea mays).
Linkage analysis and association mapping are two commonly used approaches to dissect the genetic architecture of complex traits (Darvasi and Shifman 2005). As complementary approaches, linkage analysis depends upon recent genetic recombination between two different plant lines (as the result of a genetic cross) to identify general regions of interest and identifies broad chromosome regions of interest with relatively low marker coverage, while association mapping offers high resolution takes advantage of historic recombination, and is performed by scanning a genome for SNPs in linkage disequilibrium with a trait of interest with very high marker coverage (Thornsberry et al. 2001; Hirschhorn and Daly 2005). NAM mapping strategy would combine the advantages of the two approaches to improve mapping resolution without requiring excessively dense marker maps.
NAM takes advantage of both historic and recent recombination events in order to have the advantages low marker density requirements, high allele richness, high mapping resolution and high statistical power, with none of the disadvantages of either linkage analysis or association mapping. In these regards, the NAM approach is similar in principle to the MAGIC lines in Arabidopsis.
NAM Population
A series of RIL populations developed by crossing of one common parent to a set of others is called as Nested Association Mapping population
Eg: In maize a NAM population was developed using 25 maize inbreds (testers) crossed to same inbred (founder) to develop around 250 lines for each of 25 RIL populations (Yu et al.2008).
Development of NAM population
The aim of NAM population is to capture the gens which control by rare allele for a target trait, the first step of NAM population development is selection of parents i.e.. a set of inbred lines (testers) and one founder, development of crosses between testers and founder line, advancement of F1 crosses to F7 (stable generation) by single seed decent method.
i) Selection of parents
The large set of testers generally diverse parental lines having different genes for important trait in particular crop are desirable. Many times tester can be popular varieties of different geographic region covering diverse genepool. The founder line for development of NAM population should be a popular entry which is extensively used in breeding programme, sequencing programme or reference variety in most of the genetic studies.
ii) Development of Crosses
The selected diverse lines will be chose as parental lines for NAM population development; each tester line will be cross to common Founder line to create F1 populations (Fig 1). The F1 plants will be self fertilized for six generation and advanced as single seed decent (SSD) method in order to create large set of homozygous recombinant inbreed lines (RILs).The total set of RILs will be consider as NAM population.
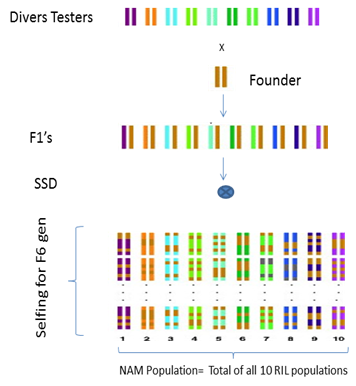
Figure 1: Procedure of development of NAM population using 10 tester lines and 1 common founder
Precautions during NAM population development
1) Maintain the pedigree of each cross while advancing to next generation.
2) The number of testers to be used for NAM population decided based on objective of work and resource (area, manpower, etc) available. Minimum 10 testers are desirable.
3) Tester used should carry different genes for important characters.
4) If complete homozygosis is not achieved in F6 generation, 1-2 generation advancement is desirable.
5) In case of cross pollinated crop avoid the contamination by foreign pollen while selfing.
Advantages of NAM population:
1) Discover the QTLs with great precision and accuracy.
2) The shuffling of many parental alleles over several generations through segregation and genetic recombination.
3) NAM population may also function as an archive for genetic diversity.
References:
1. Darvasi, A., & Shifman, S. (2005). The beauty of admixture. Nature genetics, 37(2), 118-119.
2. Thornsberry, J. M., Goodman, M. M., Doebley, J., Kresovich, S., Nielsen, D., & Buckler IV, E. S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nature genetics, 28(3), 286
3. Hirschhorn, J. N., & Daly, M. J. (2005). Genome-wide association studies for common diseases and complex traits. Nature reviews. Genetics, 6(2), 95.
4. Yu, J., Holland, J. B., McMullen, M. D., & Buckler, E. S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics, 178(1), 539-551.
About Author / Additional Info:
I am currently working as scientist in Indian Council of Agricultural Research, I have work on QTL mapping in sorghum using RIL population and presently I am developing mapping population for durum Wheat.