The local name for Boerhaavia diffusa is Punarnava which is utilized in Ayurvedic and Unani medicine. To elevate the biosafety and efficacy of the drug prepared from B.diffusa, its collection and identification is very much essential. A technique called DNA Barcoding is applied to discriminate B.diffusa from its other species. The plant is consumed in the treatment of several liver disorders and jaundice. It is used as carminative and possesses diuretic, anti-convulsant, anti-inflammatory, and fibrinolytic properties. B.diffusa is found to be contributing two pharmaceutical key alkaloids called Punarnavine-1 and Punarnavine-2. These compounds are known to be grouped under quinolizidines.
Adulteration of B.diffusa with other species of Boerhaavia like B.erecta, B.coccinea, B.repanda and B.verticillata are commonly observed. The plant B.diffusa is found to be distinct taxonomically from the other species like B.repens, B.coccinea, B.albiflora and B.tetranda. Therefore, it is not an easy job to discriminate B.diffusa from the above other species of Boerhaavia. Boerhaavia verticillata is found to be similar to B.diffusa in morphology and phytochemistry, although they differ in the plant habit.
The plant specimen can be established using DNA barcodes which is employed as a reliable, simple and efficient tool of Pharmacognosy. DNA barcodes are used to wipe out the misunderstandings in the identification of plant morphology. As the evolutionary rate is different in different taxonomic levels, nuclear ribosomal internal transcribed spacer (ITS) region is considered as a usual marker in all the evolutionary research studies. An intergenic spacer, psb A-trnH present in the chloroplast is reported to be identified in the species of Dendrobium belonging to Chinese pharmacopoeia. Hence, psb A-trnH is considered as an ultimate DNA barcode identifier.
It has been discovered in a recent study that variations in the sequence will help in constructing a specific marker for the authentication and identification of drug and herbal preparations. The current research study will help in evaluating the ideal DNA barcode identifier used for discriminating the B.diffusa from its other species, used in adulteration.
Methodology
The four species of Boerhaavia are collected from the western ghat regions at Coimbatore, India. The genomic DNA was isolated from the fresh leaves of all these plants by using the CTAB method. The amplification of ITS gene and psb A-trn H gene is performed in the presence of Taq polymerase using the reverse and forward primers of these two genes. The amplified products are collected and subjected to electrophoresis. Then the genes of ITS and psb A-trn H are purified and sequenced. The sequences of both the genes are later stored in GenBank database of NCBI. Further, the gene sequences are subjected to sequence alignment using Clustal W and MULTIALIGN web interfaces. The sequence is identified at the species level with the help of BLAST run using these two sequences. Phylogenetic trees were built using MEGA 4.0. The intraspecific differentiation among the various species was measured using MEGA 4.0. To evaluate the Wilcoxon signed rank, StatsDirect was utilized to carry out the calculations easily.
Results of the study
The genomic DNA isolated from B.diffusa, B.erecta, B.repanda and B.verticillata was used for amplifying ITS and psb A-trn H genes with PCR method. The amplified sequences were submitted to GenBank. The accession numbers of the sequences of these two genes from GenBank constitute randomly HQ386701, HQ386696, HQ407399, HQ386695, HQ386689, JF423303, HQ386691, and HQ386690. The ITS gene sequences of 11 Boerhaavia species were collected from GenBank and were utilized for sequence alignments.
The sequences of nuclear ITS gene and psb A-trn H gene of chloroplast were compared using multiple sequence alignment and pairwise alignment. The ITS region constitutes ITS1, ITS2 and 5.8S rDNA. The ITS1 sequence showed more variation compared to that of ITS2 when they were subjected to multiple sequence alignment.
Phylogenetic analysis of Boerhaavia species using ITS1 and ITS2 sequences showed that B.diffusa and B.erecta were monophyletic. When the sequence of psb A-trn H was used for the analysis B.diffusa and B.verticillata were emerged as monophyletic.
The below diagram describes the phylogenetic trees of the four Boerhaavia species constructed using ITS1, ITS2 and psbA-trnH.

Another phylogenetic tree was also created using ITS1 sequence which discriminated B.diffusa from 14 Boerhaavia species. The below phylogenetic tree shows the way in which B.diffusa differs from 14 of its other species.

The variations in nucleotides of the two genes ITS and psb A-trn H were analyzed among several species of Boerhaavia. The variation percentage shown in the figure below indicates that ITS exhibits greater inter-specific divergence.
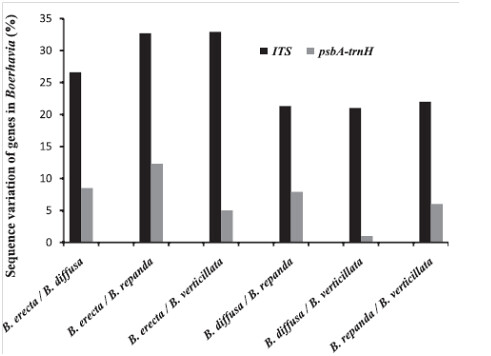
Statistical Analysis
The statistical test called Wilcoxon rank test was performed to determine the variation extent of ITS1 when compared to ITS2 and psb A-trn H genes. The statistical test results indicate that there was significant variation in the ITS1 sequence compared to the other two genes across several species of Boerhaavia dealt in this study. Sequence analysis methods like BLAST1 and Distance were also performed to indicate that ITS1 shows a greater percentage of identification at the species level.
Reference:
Dhivya Selvaraj, Dhivya Shanmughanandhan, Rajeev kumar sarma, Jijo C Joseph, Ramachandran V Srinivasan, Sathishkumar Ramalingam. DNA Barcode ITS effectively distinguishes the medicinal plant Boerhaavia diffusa from its adulterants. Genomics Proteomics Bioinformatics 10 (2012), 364-367.
About Author / Additional Info: