Author: Ashish G Vala1*, Bhupendra Singh Punwar2 and Ravi J Shah3
1Anand Agriculture University, Anand
2 National Research Center on Plant Biotechnology, New Delhi
3Department of Agriculture and cooperation, Krushi Bahvan, Gandhinagar
The development of genetic engineering has led to the creation of transgenic crops. The directed desirable gene transfer from one organism to another and the subsequent stable integration and expression of a foreign gene into the genome called genetic transformation. Selectable marker genes are widely used for the efficient transformation of crop plants. In most cases, selection is based on antibiotic or herbicide resistance. A main reason for using the marker genes in the development of transgenics is to distinguish the transformed cells from that of non-transformed cells. To produce transgenic plants, selection systems are used that lead to the selective growth of transformed cells. Genes encoding for resistance to specific antibiotics or herbicides have been found to be particularly effective for selection and provide a means for rapidly identifying transformed cells, tissues, and regenerated shoots. However, because SMGs [selectable marker gene] are integrated into the plant genome, there are concerns about widespread occurrence of transgenes in novel ecosystems (e.g., antibiotic resistance in crops and their agro ecosystems). Gene transfer from plants to environmental or medically related bacteria, or from plant products consumed as food to intestinal microorganisms or human cells, are generally considered to be not likely, but the inherent risks have not been totally addressed, and therefore there remain both regulatory and public concerns in many places in the world.
Marker gene: Powerful tool for identification of transformation events before the gene of interest can be identified in the culture. Gene transfer or DNA uptake refers to the process that moves a specific piece of DNA into cells.
"The directed desirable gene transfer from one organism to another and the subsequent stable integration and expression of a foreign gene into the genome"
Designing Genes for Insertion
Once a gene has been isolated and cloned (amplified in a bacterial vector), it must undergo several modifications before it can be effectively inserted into a plant. Simplified representation of a constructed transgene, containing necessary components for successful integration and expression.
1. Reporter or scoreable genes
• Opine synthase
• chloramphenicol acetyl transferase
• Beta Glucuronidase (GUS)
• Bacterial luciferase (Lux F2)
• The firefly luciferase (Luc)
• Green flurescent protein (GFP)
• Anthocyanins
2. Selectable markers (Antibiotic resistance markers, Antimetabolite marker, Herbicide resistance markers)
(a) Antibiotic resistance markers
• Neomycin phosphotransferase II (NPT II)
• Hygromycin phosphotransferase
• Gentamycin acetyltransferase
• Streptomycin and spectinomycin resistance
b) Antimetabolite marker
• Methotrexate-insensitive dihydrofolate reductase (dhfr)
c) Herbicide resistance markers
• Bar gene
• Enolpyruvylshikimate phosphate (EPSP) synthase (aro A) gene
• Acetolactate synthase (als) gene
• Bromoxynil nitrilase (bxn)
Marker free transgenic plants: Excise or segregate marker genes from the host genome after regeneration of transgenic plants based on the strategy called marker-free transformation.
These include methods for the removal of nuclear marker genes by intrachromosomal recombination, or using inducible hetrologous recombinases, in addition to that other novel methods for the removal of chloroplast marker genes.
Strategies to achieve markers free plants
• Co-transformation
• Site specific recombinase mediated marker deletion
• Transposons based system
• Intr-achromosomal recombination
• Removal of chloroplast marker genes
Co-tranformation : Co-transformation is a method for production of marker free transformants based on Agrobacterium- or biolistics mediated transformation in which a Selectable Marker Gene (SMG) and gene of interest are on separate constructs.
Three approaches are used for co-transformation
1. Introduction of two T-DNAs, in separate Agrobacterium strains or biolistics introduction of two plasmids in the same tissue.
2. Introduction of two T-DNAs carried by different replicons within the same Agrobacterium strain.
3. Introduction of two T-DNAs located on the same replicon within an Agrobacterium .
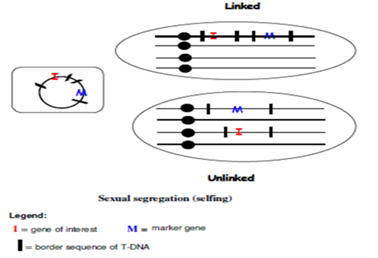
Figure 1: Linked and unlinked co- integrated genes in Agrobacterium- mediated transformation with multiple T-DNAs
Site specific recombinase mediated marker deletion: Recombination is a universal phenomenon that can occur at any place along two homologous DNA molecules. There are three well-described site-specific recombination systems that might be useful for the production of marker-free transgenic plants: Cre/loxP system from bacteriophage P1, where the Cre enzyme recognizes its specific target sites, FLP/FRT recombination system from Saccharomyces cerevisiae, where the FLP recombinase acts on the FRT sites and R/RS recombination system from Zygosaccharomyces rouxii, where R and RS are the recombinase and recombination site, respectively. The ability of site-specific microbial recombinases to cleave DNA at specific sites and ligate it to DNA at a second target sequence.
There are three site specific recombination systems..
1. Cre / lox P system from bacterophage
2. FLP/ FRT recombination system
3. R/RS recombination system
4. The simple use of microbial recombinases such as Cre , FLP and R
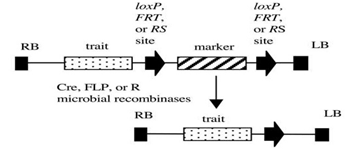
Figure 2: Site-specific recombination system (Cre/ lox) of removing a selectable marker gene
Transposon based system: Transposons are DNA sequences between hundreds to thousands of bases long. They code at least one protein, which enables them to replicate. Use of transposable elements for marker gene removal involves several steps:
(i) Insertion of the marker gene onto a transposon, a segment of DNA that “hops” around in the plant’s genome;
(ii) Co-transformation with gene of interest; and
(iii) Segregation of the marker gene.
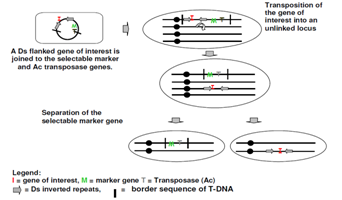
Figure 3: A Ds flanked gene of interest is joined to the selectable marker and Ac transposons genes.
MAT vector system
• MAT (multi auto transformation) vector system represent removal of nuclear marker genes.
• A chosen trait transgene is placed adjacent to a multigenic element flanked by RS recombination sites.
• Selectable ipt gene inserted between recombinase sites, together with the yeast R recombinase gene
• Entire assembly is situated within T-DNA elements.
Conclusion
Selectable marker gens have been extraordinarily useful in enabling plant transformation because of the low efficiency of transgene integration. The most used SMGs encode proteins resistant to antibiotics or herbicides and use negative selection , i.e., by killing non transgenic tissue. Transformation using marker not based on antibiotic or herbicide resistance gene, as well as different system of marker gene deletion strategies implemented. These strategies in crops and their further improvement will help to expedite widespread public acceptance of agricultural biotechnology.
Future perspectives
The efficient and practical approaches are still needed in order to introduce multiple genes into plant genomes. Transformation efficiencies can be enhanced to a level at which the use of selectable markers becomes unnecessary. Improvement of existing strategies and the development of novel approaches for plant genome manipulation are required.
References:
1. Abolade S. (2007). Status of clean gene (selection marker free) technology, African journal of biotech, 6[25], 2910-2923.
2. Behrooz D, Amin E, Neal stewart C, and Willam N. (2007). Methods to produce marker free transgenic plants. Biotechnol. J. [2];83-90.
3. Caharles P, Elena Z, Peter M. (2002). Techniques for the removal of marker genes from transgenic plants. Biochimie, 1119-1126.
4. Cox M. M.(1989). DNA inversion in the 2 um plasmid of Saccharomyces cerevisiae, in: Berg, D. E., Howe, M. M.(Eds.), Mobile DNA, American Society for Microbiology, Washington D.C.661-670.
5. Erikson O, Hertzberg M, Nasholm T. (2004). A conditional marker gene allowing both b positive and negative selection in plants. Nat. Biotechnol.,[ 22], 455-458.
6. Hartl D L, Jones E W. (1998). Genetics: principles and analysis, 4th edn., London, pp. 330-359.
7. Hoess R H. and Abremski K. (1990). The Cre/lox recombination system. Nucleic Acids Mol. Biol.,[4], 99-109.
8. Huang L C, Wood E A. and Cox M M. (1991). A bacterial model system for chromosomal targeting. Nucleic Acids Res. [19], 443-448.
9. Jhon I. and Andrew P. (1994). Transformation systems for generating marker free transgenic plants., Nucleic Acids Res., [12], 263-267.
10. Iamtham S. and Day A.(2000). Removal of antibiotic resistance genes from transgenic
11. tobacco plastids. Nat. Biotechnol., [18], 1172-1176.
12. Joersbo M, Donaldson I, Kreiberg J. and Petersen S G. (1998). Analysis of mannose selection used for transformation of sugar beet. Mol. Breed., [4], 111-117.
13. Kerry A L, Massimo H B. and Pal M.(2006) Plastid marker-gene excision by transiently expressed CRE recombinase. Plant J., [45] 447.
14. Kumar S, Arul L. and Talwar D. (2010). Generation of marker free Bt transgenic indica rice and evaluation of its yellow stem borer resistance. J. Appl Genet., 51[3], 243-257.
15. Khan R S, Nakamura I. and Mii M. (2010). Production and selection of marker free transgenic plants of Petunia hybrid using site specific recombination. Biologica Plantarum., 54[2], 265-271.
16. Lucca P, Ye X. and Potrykus I.(2001). Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol. Breed., 7, 43-49.
17. Lu Lu X W, Xiaoyan Y, Jonathan M, Xinlu C, William R F. and Zhanyyuan J Z. (2009). development of marker- free transgenic sorghum [Sorghum bicolor (L.) Moench] using standard binary vectors with bar as a selectable marker, Plant Cell Tiss Organ Cult., 99,97-108.
18. Matsuzaki H, Nakajima R, Nishiyama J. and Araki H. (1990). Chromosome engineering in Saccharomyces cerevisiae by using site-specific recombination system of a yeast plasmid. J. Bacteriol., [172], 610-618.
19. Negrotto D, Jolly M, Beer S. and Wenck A R. (2000). The use of phosphomannose- isomerase as a selectablemarker to recover transgenic maize plants (Zea mays L.) via Agrobacterium transformation. Plant Cell Rep.798-803.
20. Okennedey M M, Burger J T. and Botha F C. (2004). Pearl millet transformation system using the positive selectable marker gene phosphomannose isomerase. Plant Cell Rep., [22], 684- 690.
21. Pawan K, Lingaraj S, Dolendro S and Rana P. (2002) Strategies to deal with the concern about marker genes in transgenic plants: some environmental friendly approaches, currentsci., [83], 2-5.
22. Reed J, Privalle L, Powell M L and Meghji M. (2001). Phosphomannose isomerase: an efficient selectable marker for plant transformation.In Vitro Cell Dev. Biol. Plant.[ 37], 127-132.
23. Ravi R, Suchandra D, Mikhali P, Thomas H and Neera B. (2007). Marker free approach for developing abiotic stress tolerant transgenic Brassica Juncea (Indian mustard). Int . conference on cellular and molecular biology, 26-28.
24. Sternberg N, Hamilton D. (1981) Bacteriophage P1 site-specific recombination between loxP sites. J. Mol. Biol., 150, 467-486.
25. Sonntag K, Wang Y. and Wallbraun M. (2004). A transformation method for obtaining marker-free plants based on phosphomannose isomerase. Acta Univ. Latviensis Biol. 676, 223-226.
26. Wright M, Dawson J, Dunder E and Suttie J.(2001). Efficient biolistic transformation of maize (Zea mays L.) using the phosphomannose isomerase gene, pmi, as the selectable marker. Plant Cell Rep., [20], 429-436.
27. Yue Z, Hua L, Bei L, Jian-Tao Z, Yizjou L. and Hongxia Z. (2009). Generation of selectable marker free transgenic tomato resistant to drought, cold and oxidative stress using the Cre/loxP DNA excision system, transgenic Res,[18], 607-619.
28. Zhu L, Fu Y P, Liu W Z. and Hu G. (2007). Rapid Generation of Selectable Marker-Free Transgenic Rice with Three Target Genes by Co-Transformation and Anther Culture. Rice Science., 14[4], 239-246.
29. Zubko E, Scutt C. and Meyer P. (2000). Intrachromosomal recombinationbetween attP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat. Biotechnol. [18], 442-445.
About Author / Additional Info:
Doctorate in Plant Biotechnology and research area is rice functional genomics.