Author: Dr. Nalin K. Pagi
Allele Mining: Searching for different alleles. Allele - alternative forms of a genetic character found at a given locus on a chromosome. Mining - Searching. It is often referred to as ‘dissection of naturally occurring variation at candidate genes/loci’ or simply ‘allele mining’.
Why Allele mining?
- Direct access to alleles conferring: resistance to biotic stresses, tolerance of abiotic stresses greater nutrient use efficiency, enhanced yield and improved quality.
- Despite the fact that the propagation of these altered/novel alleles (either through natural or artificial selection) depends upon their role in the plant's adaptation, it can be suggested that such altered/novel alleles of key genes are preserved in many related wild species, land races and cultivated varieties of a crop species.
- Therefore, exploring the natural variation in important genes is highly indispensable in modern crop improvement programs.
- Two major approaches are available for the identification of sequence polymorphisms for a given gene in the naturally occurring populations.
- Modified TILLING (Targeting Induced Local Lesions in Genomes) procedure called EcoTILLING and
- Sequencing based allele mining.
- A brief description of these two approaches is given in this section and steps involved in these approaches are illustrated in Figure 1.
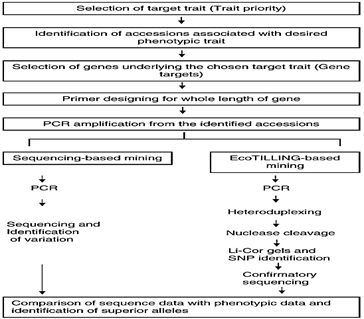
- TILLING is a technique that can identify polymorphisms (more specifically point mutations) resulting from induced mutations in a target gene by heteroduplex analysis (Till et al., 2003).
- A variation of this technique, Eco TILLING, represents a means to determine the extent of natural variation in selected genes in crops. The method is essentially same as TILLING except that the mutations are not induced artificially and are detected from naturally occurring alleles in the primary and secondary crop gene pools (Comai et al., 2004).
- Like TILLING, EcoTILLING also relies on the enzymatic cleavage of heteroduplexed DNA (formed due to single nucleotide mismatch in sequence between reference and test genotype) with a single strand specific nuclease (i.e., Cel-1, Mung bean nuclease and S1 nuclease etc.) under specific conditions followed by detection through Li-Cor genotypers (Li-Cor, USA).
(Ashkani et al., 2015)
Sequencing-based allele mining:
- Sequencing-based allele mining involves PCR-based amplification of alleles of a gene in varied genotypes and then DNA sequencing to recognize nucleotide variance in the alleles. Various alleles among the cultivars through this approach can be identified.
- The method would help to analyse individuals for haplotype structure and diversity to infer genetic association studies in plants.
- Using this approach to recognize the effect of mutations on gene structure, the sequences are analysed for the location of point mutations or SNPs and insertions or deletions (InDels) to construct haplotypes.
- Additionally, this method is applicable to the management of additional crop diseases. Sequencing-based allele mining does not require much sophisticated equipment or involve tedious steps (Ramkumar et al., 2010).
Application of allele mining:
- Allele mining can be effectively used for discovery of superior alleles, through ‘mining’ the gene of interest from diverse genetic resources. It can also provide insight into molecular basis of novel trait variations and identify the nucleotide sequence changes associated with superior alleles.
- In addition, the rate of evolution of alleles; allelic similarity/dissimilarity at a candidate gene and allelic synteny with other members of the family can also be studied.
- Allele mining may also pave way for molecular discrimination among related species, development of allele-specific molecular markers, facilitating introgression of novel alleles through MAS or deployment through genetic engineering (GE).
Prospects:
- Allele mining can be visualized as a vital link between effective utilization of genetic and genomic resources in genomics-driven modern plant breeding.
- Firstly, there is a need to develop more refined sampling strategies or tools which can aid in such selection to enrich useful alleles in a minimum number of genotypes with desired levels of trait expression.
- Secondly, to effectively extract and apply information from large genomic resources, several molecular, biological and bioinformatic tools are currently available.
- The outcome of international genome sequencing projects will make allele mining possible for many genes in different crop species and possibly from related species within the limits of genome synteny and sequence conservation.
- As we are entering an era of low cost and high throughput sequencing, routine allele mining through sequencing would soon become a universal tool to score variations at candidate genes between individuals and become an effective as well as economically viable method of choice for the identification and development of novel genetic resources for deployment in plant breeding.
References:
Ashkani, S.; Yusop, M. R.; Shabanimofrad, M.; Azadi, A.; Ghasemzadeh, A.; Azizi, P. and Latif, M.A. (2015). Allele Mining Strategies: Principles and Utilisation for Blast Resistance Genes in Rice (Oryza sativa L.). Curr. Issues Mol. Biol., 17: 57-74.
Comai, L., Young, K., Till, B.J., Reynolds, S.H., Greene, E.A., Codomo, C.A., Enns, L.C., Johnson, J.E., Burtner, C. and Odden, A.R., 2004. Efficient discovery of DNA Curr. Issues Mol. Biol. 17:57-74. horizonpress.com/cimb 68 Allele Mining for Blast Resistance in Rice Ashkani et al. polymorphisms in natural populations by Ecotilling. The Plant Journal 37: 778-786.
Ram Kumar, G.; Sakthivel, K.; Sundaram, R.M.; Neeraja, C.N.; Balachandran, S.M.; Shobha Rani, N.; Viraktamath, B.C. and Madhav, M.S. (2010). Allele mining in crops: Prospects and potentials. Biotechnology Advances, 28: 451-461.
Till, B.J., Reynolds, S.H., Greene, E.A., Codomo, C.A., Enns, L.C., Johnson, J.E., Burtner, C., Odden, A.R., Young, K. and Taylor, N.E., 2003. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Research. 13: 524-530.
About Author / Additional Info:
I am currently working as a Senior Research Fellow at S.D. Agricultural University. I have done Ph.D. in Genetics and Plant Breeding and M.Sc. in Botany. I have also worked with S.D.Agricultural University as a Visiting Lecturer for 2 years.