Author: Anupama Singh
The major challenge in transgenic plant development is to generate plants with stably integrated transgenes over generations. Here, some methods are discussed for study of transgene integration, location and its expression.
- PCR (Polymerase chain reaction)
PCR variants :-
- Real time quantitative PCR It is used for the rapid estimation of transgene copy number in comparison with control sample having a single copy endogenous gene to be amplified. The transgene copy numbers are revealed by relative signal intensities of the control and the transgenic plants. It also shows presence/ absence of transgene and provide information of copy number estimate, and a little way to information about structure of transgene locus with the genomic flanking sequences already known.
- Inverse PCR, TAIL-PCR and plasmid rescue Transgenic locus identified by these PCR variants followed by DNA sequencing reveals the precise structural organization and rearrangement, the nature of transgene genomic and transgene- transgene junctions at the nucleotide level and integration site in genomic DNA.
- Southern blotting The technique used in analysis of transgenic loci. It provide different information depending on the restriction enzyme and probes used, different sized fragments produced depending on fixed point in transgene and the adjacent restriction site in genomic DNA which varies according to site of insertion. It also provides estimate about transgene copy number. The vector sequence are also used as probe that will hybridize to vector backbone which identifies the vector contamination in the transgenic plant.(Fig 1)
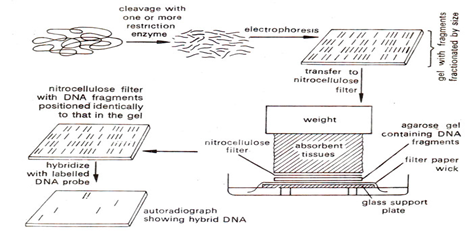
Fig 1:- Steps in southern hybridization
- Fibre-FISH( Fluorescent in-situ hybridization) It allows the characterization of the transgenic loci at chromosomal level by using isolated DNA fibres, interphase chromatin and metaphase chromosome. The fluorescently labelled target sequence probes are used to identify particular target sequence that reveals distribution of integration sites and higher order transgene organization. FISH using metaphase chromosome allows mapping of insertion sites cytogenetically and in interphase determination of nuclear territory of transgene. FISH on extended chromatin provides an overview of locus structure, presence of single copy inserts, interspersed genomic DNA and transgene concatemers. The FISH resolution lies between Southern hybridization and genetic segregation and provide important correlative data for both the techniques.
- Segregation analysis It involves the study of transmission of particular DNA sequence over several generation of transgenic plants. It allows determination of number of transgenic loci on the basis of Southern blot hybridization rather than phenotype analysis ( includes misleading results caused by epigenetic gene silencing).
- Next generation sequencing Whole genome sequencing and targeted capture sequencing gives information about transgene copy number, T-DNA integrity, presence/ absence of vector backbone sequences and stability of T-DNA inserts across different generations. It is highly sensitive, cost and labor effective as compared to traditional southern blot analysis.(Fig 2)
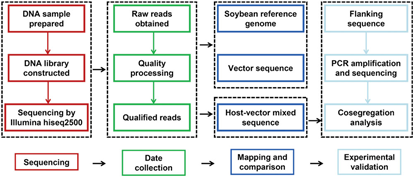
Fig 2:- Schematic flow chart for insertion site identification and validation
- Guo Bingfu, Guo Yong, Hong Huilong, Qiu Li-Juan, Identification of Genomic Insertion and Flanking Sequence of G2-EPSPS and GAT Transgenes in Soybean Using Whole Genome Sequencing Method
About Author / Additional Info:
Ph.D scholar at National Research Centre on Plant Biotechnology, New delhi