Author: Lalbahadur Singh
Introduction
Mature miRNAs are single-stranded short RNA sequence with 21–24 nucleotides in length that base pair with target mRNAs. Generally, the miRNA biogenesis involves several interdependent steps, including primary miRNAs (pri-miRNAs) transcription, further processing and modification, RNA induced silencing complex (RISC) loading. The transcription of plant pri-miRNAs is similar to protein coding genes, the majority of them are transcribed from their own transcriptional units termed as MIR genes, whose genome sequences are usually located at intergenic regions of protein coding genes and have their own promoter and independent regulatory pattern. Prior to further processing, pri-miRNAs will be capped at their 5’ end and polyadenylated at their 3’ end similar as the most other pol II derived transcriptional event. In addition, pri-miRNAs contain short ORF (open reading frame) sequences involved in the synthesis of regulatory peptides, which can promote the accumulation of their own mature miRNAs.
miRNAs Role in Plant Morphogenesis
Plant stem cells are confined within specialized niches, shoot apical meristems (SAM), and root apical meristems (RAM), respectively. In higher plants, the formation of shoot meristems generally occurs in two situations, first SAM formation occurs during embryogenesis from the axil of cotyledon(s), and the second SAM formation occurs during post-embryogenesis from the axil of leaves. As the critical role of stem cells in plant morphogenesis, meristems studies are always the hot research area. miRNA play role in growth and development of different plant part as described in fig 1.
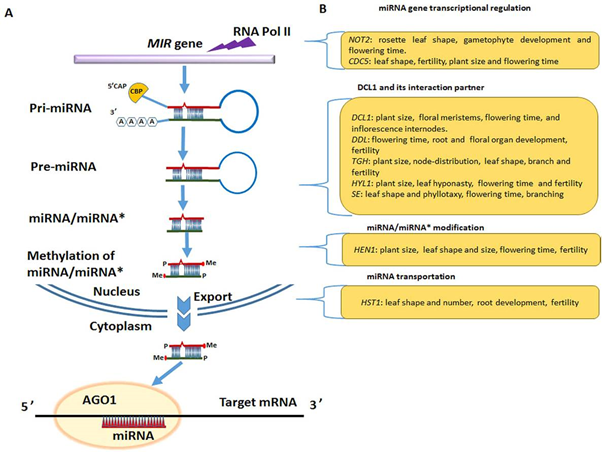
Fig 1 Schematic model of stem cell-derived plant organogenesis and miRNA-mediated regulation. (Weigel and Jurgens, 2002).
Impairment of miRNA Biogenesis Genes Cause Pleiotropic Developmental Defects
In plant, the DNA-dependent transcriptional activator, RNA polymerase II (Pol II), is majorly responsible for transcribing those MIR genes to generate large initial transcriptional product referred as primary miRNAs. During this step, some auxiliary proteins, such as mediator complex, Negative on TATA less2 (NOT2) protein, elongator complex, assist Pol II to increase its activation.
pri-miRNAs harbor an imperfect stem-loop structure, which is need for directing DICER-LIKE1 (DCL1)-mediated cleavage near the base of its stem to generate a precursor miRNA (pre-miRNA). And the pre-miRNA stem-loop structure is further processed by DCL1 into miRNA/miRNA_ duplex. For stabilization, the 3’ end of miRNA/miRNA* duplex is methylated by RNA methyltransferase HUA ENHANCER 1 (HEN1) in the nucleus (Kurihara and Watanabe, 2004). In Arabidopsis, argonaute protein 1 (AGO1), which possesses endonuclease activity, is the major effector that is responsible for recruiting miRNA to form RISCs. In this AGO1 centered RISCs, the mature single stranded miRNA functioned as guides to target complementary mRNAs, while the miRNA* (passenger strand) is often destabilized.
Intriguingly, intensive studies indicate that plant miRNAs biogenesis are tightly regulated, otherwise plants would exhibit multiple developmental defects, such as plant size, flowering time, and fertility (Fig.1), suggesting that miRNAs are the potent regulators in plant development.
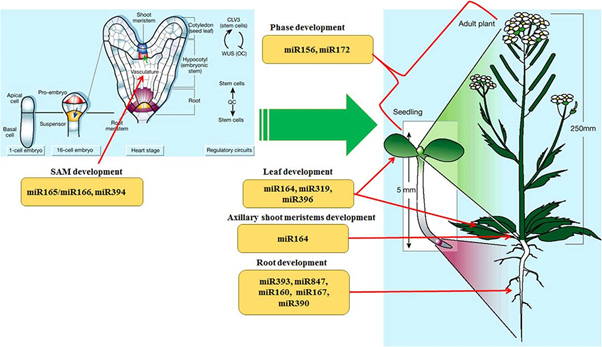
Fig 2 Misregulation of miRNA biogenesis results in pleiotropic developmental defects. A: A diagrammatic representation of miRNA biogenesis processes. B: Severe developmental impairments result from the mutation of key miRNA biogenesis genes. Abbreviation- Negative on TATA less2 (NOT2), CELL DIVISION CYCLE 5 (CDC5), DICER-LIKE1 (DCL1), DAWDLE (DDL), TOUGH (TGH), HYPONASTIC LEAVES1 (HYL1), SERRATE (SE), HUA ENHANCER 1 (HEN1), HASTY 1 (HST1).
Conclusion
Based on the intensive researches in the past 20 years, rapid and significant progress has been made in uncovering miRNA biogenesis, targets prediction, biological functions, and molecular mechanisms, which greatly advance our understanding about the elaborate and fancy regulatory network generated with long-term plant evolution. In addition, it is a tendency that miRNA can act as a new breeding tool in plant genetic improvement. However, the majority of the miRNA are not well studied, especially some of low-abundance miRNAs, which also exert potent role in plant development as described recently. Additionally, comparing with multifunctional and dominant roles, miRNAs still have many mysterious areas need to be further studied.
References:
- Weigel, D., Jurgens, G. (2002). Stem cells that make stems. Nature, 415, 751–754.
- Lauressergues, D., Couzigou, J. M., Clemente, H.S, Martinez, Y., Dunand, C., Becard, G., Combier, J.P. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature, 520, 90–93.
- Li, C., Zhang, B. (2016). MicroRNAs in Control of Plant Development. J. Cell. Physiol., 231, 303–313.
About Author / Additional Info:
Ph.D. scholar, biotechnology, Indian agricultural research institute (IARI), new delhi. Currently I am working in the area of miRNAs in pulse crop.