Authors: A. Kandan, J. Akhtar, Pardeep Kumar and Z. Khan
Division of Plant Quarantine, ICAR-National Bureau of Plant Genetic Resources (NBPGR), Pusa Campus, New Delhi â€" 110012
Proteomic analysis may be defined as the global assessment of cellular proteins expressed in a particular biological state, which is a powerful tool that can provide a systematic understanding of particular events at the molecular level. Here, in-depth studies of plantâ€"pathogen interactions using the proteomic approach begun to be applied in plant pathology field. Invention and usage of mass spectrometry technology, different protein stains, software and good progress in bioinformatics have made identification of proteins relatively easy. Since last two decades 2D gel electrophoresis has been used as the primary tool for obtaining a global picture of the expression profile of a proteome under various conditions either plant tissue or pathogen or both. In this method, first proteins are first separated in one direction by isoelectric focusing and later in the orthogonal direction by molecular mass using electrophoresis in a slab gel containing SDS-PAGE. In recent years, these protein separation methods coupled with various MS technologies have evolved as the dominant tools in the field of biology as protein identification and protein complex deconvolution. In MS/MS analysis, a particular ion is first selected with a mass filter and then the selected ion is further fragmented and analyzed. This Fragmentation can be first induced by introducing the ion into a protein chamber with a collision gas such as argon where collision of ions and argon atoms results in fragmentation. The daughter ion spectrum is then analyzed and documented. Further several uses of bioinformatic tools to identify the protein from their respective database (Fig. 1).
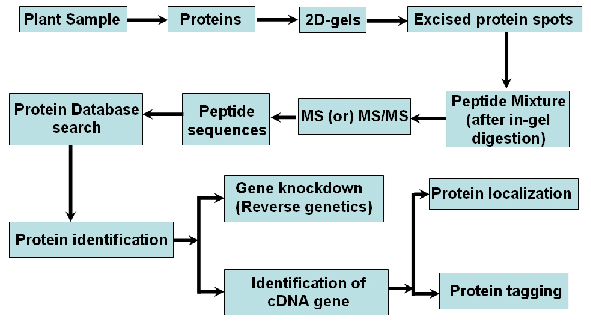
Fig. 1. Schematic diagram to show the principles of proteomics in host-pathogen interaction studies.
Plant-fungus interactions
Proteomic studies addressing the plant-fungus interactions are as follows: A basidiomycete biotrophic rust fungus Puccinia triticina challenge inoculated in both susceptible and resistant wheat cultivar showed no relevant difference between inoculated and mock inoculated plants in resistant cultivar, whereas susceptible cultivar showed up regulation of proteins which mainly involved in the metabolic enzymes, structural proteins, and proteins with a role in pathogenesis. Proteins associated with cell defense, metabolism and cell differentiation showed altered abundance compared to non-infected controls when Arabidopsis thaliana inoculated with Plasmodiophora brassicae. When maize seed embryos are germinated in the presence of the seed borne fungus Fusarium verticillioides, it was clearly observed that several proteins were significantly up regulated viz. Pathogenesis Related (PR) proteins, antioxidant enzymes and several other proteins involved in protein synthesis, protein folding and stabilization. In addition, increase in protein levels of a fructose-bisphosphate aldolase protein (cytoplasmic isoform), while other enzyme mainly involved in carbon metabolism, like glyceraldehyde 3-phosphate dehydrogenase, was effectively down-regulated in response to Fusarium verticillioides fungal infection.
Plant-bacteria interactions
Several scientists already reported the interactions of various phytopathogenic bacteria with their respective hosts through different proteomic approaches. Modification of phosphorylation proteins which eventually trigger the signaling cascades of host against the pathogens. Both mitogen activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs) can play major role in pathogen recognition and in the downstream events that eventually lead to plant defense mechanism. Two induced antioxidant enzyme groups (glutathione S-transferases and peroxiredoxins) observed in the soluble fraction of Arabidopsis leaf proteins following Pseudomonas syringae pathogen challenge inoculation. Scientists also observed three subcellular fractions (total soluble protein, chloroplast-enriched protein, and mitochondria-enriched protein) over a 1.5 - 6 h time period following Pseudomonas syringae inoculation. Interestingly in both studies differential induction of defense-related antioxidants and various metabolic enzymes observed as the main changes in protein expression identified.
Researchers interested to identify bacterial blight (Xanthomonas oryzae pv. oryzae) pathogen defense-responsive proteins in rice, first either leaf blades from rice seedlings or suspension cells of different rice cultivars were treated with both their compatible and incompatible Xanthomonas oryzae pv. oryzae races. Several proteins from rice plant cytosolic and/or membrane fractions were differentially well expressed in response to Xanthomonas oryzae pv. oryzae bacterial inoculation and observed some of these proteins were known to function in plant defense. Both common and dissimilar patterns of protein expression changes were observed between the compatible and incompatible interactions of rice host-bacterial interactions (Chen et al. 2007). Tomato (Lycopersicon hirsutum) lines carrying two quantitative trait loci viz. Rcm 2.0 and Rcm 5.1 and susceptible control plants challenge inoculated with bacteria (Clavibacter michiganensis subsp. michiganensis) causing canker of tomato showed oxidative stress-related proteins in response to pathogenic bacterial infection, and distinct protein regulation patterns by the tomato plant lines carrying the Rcm 2.0 and Rcm 5.1 mutations were revealed, suggesting that these tomato lines they may confer resistance via independent pathways.
A differential multiplexed proteomic analysis identified a huge spectrum of differentially expressed Nicotiana tabacum proteins in response to lipopolysaccharide (LPS) of bacteria Burkholderia cepacia, which is a plant immunity elicitor extracted from an endophytic strain of Burkholderia cepacia. Further, functional classification of these spectrum of proteins contributed to a better understanding of the effective elicitation mechanism of this host defense responses and led to the conclusion that the overall effects of LPSB.cep on Nicotiana tabacum cells were the effective induction of metabolic reprogramming and appreciable changes in cellular activities supportive of protein synthesis, degradation, folding and as well as vesicle trafficking and their secretion and also cytoskeleton transformation (Gerber et al. 2008).
Few researchers reported the plant-symbiotic relation with antagonistic microorganisms through proteomic approaches. Several proteins that were differentially regulated during the symbiotic interaction of root colonizing microbes with Medicago truncatula host and observed new proteins of unknown function as well as confirmed known symbiosis related proteins. Few proteins thought to be expressed only under conditions of phytopathogenic attack were also identified including glutathione-S-transferase (GST), peroxidase, and polygalacturonase inhibitor protein (PGIP). Some other proteins were involved in plant development regulation or nitrogen fixation for effective growth of the host. Another interesting work on three-way interaction involved bean plants, a fungal pathogen (Botrytis cinerea) and an antagonistic strain of the fungus Trichoderma atrviridae. The proteomes secretions of the partners involved were analyzed either alone, in all combinations of two and the complete interaction involving the three partners. This interesting experimental set up permitted the host-pathogen-antagonist partners to interact with each other as the cellophane membrane allowed the diffusion of micro- and macro-molecules, at the same time these partners could be separated with each other and their proteins extracted from each other separately showed interesting homologies with a fungal hydrophobin, ABC transporters, etc. Few virulence factors, like cyclophilins, were up-regulated in the pathogen proteome secretion during the interactions with the plant alone or with the antagonist too. Thus proteomic approaches allowed us to identify several differential proteins involved in cross-talk normally occurring in nature between hosts, pathogens and various biocontrol agents.
References:
1. Chen, F., Yuan, Y., Li, Q., He, Z., 2007. Proteomic analysis of rice plasma membrane reveals proteins involved in early defense response to bacterial blight. Proteomics 7: 1529-1539.
2. Gerber, I.B., Laukens, K., de Vijlder, T., Witters, E., Dubery, I.A., 2008. Proteomic profiling of cellular targets of lipopolysaccharide-induced signaling in Nicotiana tabacum BY-2 cells. Biochimica et Biophysica Acta 1784: 1750-1762.
About Author / Additional Info:
Senior Scientist (Plant Pathology), Division of Plant Quarantine, Indian Council of Agricultural Research (ICAR)-National Bureau of Plant Genetic Resources (NBPGR), Pusa Campus, New Delhi-110012.