Authors: Pooja Kumari*, Raj Kiran* and Atul Kumar**
* Scientist , Division of plant Quarantine, ICAR-National Bureau of Plant Genetic Resources, New DELHI-110012
** Principal Scientist, Division of Seed Science and Technology, ICAR-Indian Agricultural Research Institute, New DELHI-110012
Siderophores (from the Greek: “iron carriers”) are defined as relatively low molecular weight, ferric ion specific chelating agents elaborated by bacteria and fungi growing under low iron stress. The role of these compounds is to scavenge iron from the environment and to make the mineral. Most organisms require iron as an essential element in a variety of metabolic and informational cellular pathways. More than 100 enzymes acting in primary and secondary metabolism possess iron-containing cofactors such as iron-sulfur clusters or heme groups. The reversible Fe(II)/Fe(III) redox pair is best suited to catalyze a broad spectrum of redox reactions and to mediate electron chain transfer. Furthermore, several transcriptional (e.g., bacterial Fur and PerR) and posttranscriptional (e.g., mammalian iron regulatory proteins [IRPs]) regulator interact with iron to sense its intracellular level or the current status of oxidative stress in order to efficiently control the expression of a broad array of genes involved mainly in iron acquisition or reactive oxygen species (ROS) protection. The cellular uptake of iron is restricted to its physiologically most relevant species, Fe(II) (ferrous iron) and Fe(III) (ferric iron). Fe(II) is soluble in aqueous solutions at neutral pH and is hence sufficiently available for living cells if the reductive state is maintained. Generally, Fe(II) can be taken up by ubiquitous divalent metal transporters. Systems for specific Fe(II) uptake are known in bacteria and yeast. Siderophore-dependent iron acquisition pathways can be found among a broad spectrum of prokaryotic and eukaryotic microbes (and even in higher plants) and show a high variety in structure and function of the involved components. The common theme is the production of one or more siderophores by cells during periods of iron starvation (which means that the intracellular iron concentration drops below the threshold of about 10-6M, which is critical for microbial growth). Secreted siderophores form extracellular Fe(III) complexes with stabilities ranging over about 30 orders of magnitude for different siderophores. Next, either the iron charged siderophore is taken up by ferric-chelate-specific transporters or siderophore-bound Fe(III) undergoes reduction to Fe(II), which is catalyzed by free extracellular or membrane- standing ferric-chelate reductases.
SIDEROPHORES IN PLANT PATHOGENIC BACTERIA
The role of microbial siderophores in virulence to plant hosts was first demonstrated for the bacterial pathogen Erwinia chysanthemi which produces the catecholate chrysobactin and the carboxylate achromobactin. Erwinia amylovora synthesizes the hydroxamate desferrioxamine and mutants defective in desferrioxamine biosynthesis show tissue-specific reduced virulence. Siderophore is not only involved in iron uptake but also play a key role to develop resistance against antibiotic.
Cellular transport systems for uptake of siderophore-delivered iron in bacteria and fungi
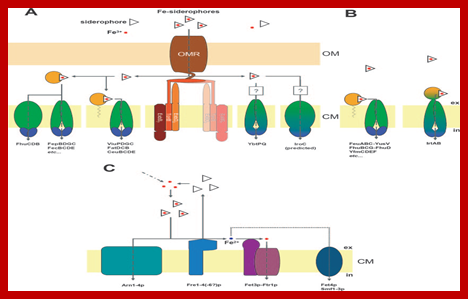
FIG. 1. Cellular transport systems for uptake of siderophore-delivered iron in bacteria and fungi. (A) Gram-negative bacteria. Fe-siderophore uptake through the OM is mediated by an OM receptor (OMR), which is energized by the proton motive force transduction system ExbB7ExbD2-TonB (“TonB complex”). The second TonB complex shown in faint colors suggests a possible 2:1 interaction with the OM receptor. Fe-siderophore uptake through the inner membrane depends on ABC-type transporters with different domain arrangements and localizations periplasmic binding proteins/domains are shown in bright orange,membrane-spanning proteins/domains are shown in green, and cytoplasmic ATP-binding proteins/domains are shown in dark blue. Domain fusions are indicated by corresponding color transitions.Question marks point to the unknown involvement of periplasmic binding proteins.(B)Gram-positive bacteria.Fe-siderophore uptake through the CM by known ABC-type transporters is shown. (C) S. cerevisiae. Fe-siderophore uptake is either mediated by MFS transporters (Arn proteins) or Fe-siderophores are reduced extra cytoplasmatically by membrane-standing metalloreductases (Fre proteins). Released ferrous iron either is then reoxidized and taken up by the multicopper ferroxidase-high-affinity uptake complex (Fet3p-Ftr1p) or may be imported by divalent metal transporters such as Smf proteins (low specificity) or Fet4p (low affinity and specificity),(Marcus Miethke and Mohamed A. Marahiel 2007).
FUNGAL IRON ACQUISITION SYSTEMS :
In fungi, four different mechanisms for iron uptake have been characterized at the molecular level: (i ) siderophore-mediated Fe3+ uptake, ( ii ) Reductive iron assimilation (RIA) (iii ) heme uptake, and (iv) direct Fe2+ uptake. (i ) and (ii ) are high-affinity iron uptake systems; ( iii ) represents the utilization of a special iron source typically found within hosts; and (iv) is a low-affinity iron acquisition system. Most fungal species employ more than one of these systems in parallel but rarely are all four strategies present in the same species. High-affinity systems are important under iron-limited conditions, whereas low affinity systems are of use only when iron is abundant. All fungal siderophores identified so far are hydroxamates. Fungal hydroxamates are derived from the nonproteinogenic amino acid ornithine and different acyl groups and can be grouped into four structural families: (i ) rhodotorulic acid, (ii ) fusarinines, (iii ) coprogens, and (iv) ferrichromes.
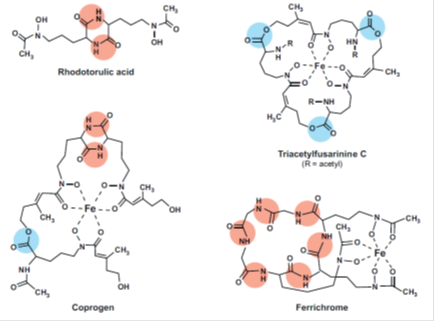
FIG. 2. Representative fungal siderophores
Fungal siderophore biosynthesis
The first committed step in the biosynthesis of fungal hydroxamate siderophores is N5-hydroxylation of ornithine catalyzed by ornithine-N5-monooxygenase. The second step in siderophore biosynthesis results in the formation of the hydroxamate group, accomplished by N5- acylation of N5-hydroxyornithine.The hydroxamate group is formed by transfer of an acyl group from acyl-coenzyme A derivatives to N5-hydroxyornithine. Here, the pathway for different siderophores splits for the first time due to the choice of the acyl group, with the simplest being acetyl (e.g., for rhodotorulic acid, FC and ferrichrome) or more complex ones such as anhydromevalonyl (e.g., for fusarinines and coprogens) or methylglutaconyl (e.g., for ferrichrome A). In the third siderophore biosynthetic step, the hydroxamates are covalently linked via ester (fusarinines, coprogens) or peptide bonds (rhodotorulic acid, ferrichromes, coprogens) accomplished by nonribosomal peptide synthetases (NRPSs). NRPSs are large multifunctional enzymes that synthesize peptides from proteinogenic and nonproteinogenic precursors independently of the ribosome. NRPSs have a modular structure. One full module harbors all the catalytic units for incorporation of one amino acid (or amino acid-like) residue: an adenylation domain (A) for substrate specificity and activation, a peptidyl carrier domain (T) for attachment of the activated substrate, and a condensation domain (C) for bond formation.
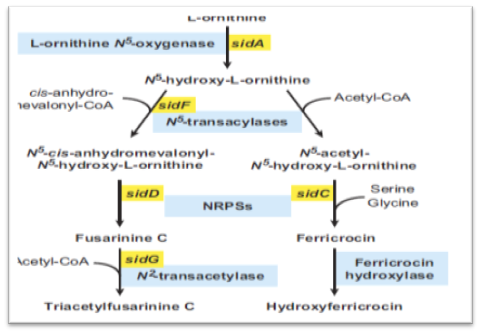
FIG. 3. Postulated siderophore biosynthetic pathway of A. fumigatus. Biosynthesis of both TAFC and FC starts with N5-hydroxylation of ornithine. Subsequently, the hydroxamate group is formed by the transfer of an acyl group from acyl-coenzyme A (CoA) derivatives to N5-hydroxyornithine. Here the pathways for biosynthesis of TAFC and FC split due to the choice of the acyl group with acetyl for FC and anhydromevalonyl for TAFC. Assembly of the cyclic siderophores fusarinine C and FC is catalysed by different non-ribosomal peptide synthetases (NRPS). TAFC and hydroxyferricrocin are formed by N2-acetylation of fusarinine C and hydroxylation of FC respectively. With exception of the acetyl transferase required for FC biosynthesis all A. fumigatus genes encoding respective enzyme activities have been identified and are indicated (Schrettl et al., 2004; 2007).
Characterized Pathogenic Fungal Siderophore NRPS
| NRPS name | Fungal species | Siderophores | Reference |
| Ferrichrome NRPS | |||
| Nps2 | C. heterostrophus | Ferricrocin | Oide et al., 2007 |
| Nps2 | F. graminearum | Ferricrocin | Oide et al., 2007 |
| SidFA/Fer3 | U. maydis | Ferrichrome A | Eichhorn et al., 2006 |
| Coprogen NRPS | |||
| Nps6 | A.brassicicola | N-dimethylcoprogen | Oide et al., 2006 |
| Nps6 | C.heterostrophus | Coprogen | Oide et al., 2006 |
| Nps6 | C.miyabeanus | - | Oide et al., 2006 |
| Fusarinine NRPS | |||
| Nps6 | F.graminearum | Fusarinine C | Oide et al., 2006 |
Application
- Siderophore helps in increased iron availability to plants.
- It helps to increase the population of beneficial root colonizing Bacteria in rhizosphere soil.
- Used as biocontrol agent
- Used as drugs in facilitating iron mobilisation in humans Conclusion In recent years it became clear that the siderophore system constitutes a central element in iron homeostasis of many if not most fungi, affecting growth, oxidative stress resistance,as well as asexual and sexual development. Moreover, it became clear that siderophores are a common virulence determinant of at least some animal- and plant-pathogenic fungal species, and that siderophores are also involved in symbiosis. The role of individual iron homeostasis-maintaining mechanisms in virulence, however, depends largely on the pathogen-host system. The fungal requirement for iron could potentially open up perspectives towards the development of novel antifungal treatments, e.g., iron chelation therapy or blocking of high-affinity iron acquisition or development of chemical surrogates for siderophores. For siderophore-synthesizing fungi, blocking of siderophore transporters or siderophore-biosynthetic enzymes appears to be particularly promising because the involved proteins are not present in mammals and plants. Recently, the first antibacterial agent targeting siderophore biosynthesis of pathogenic bacteria have been reported. Apart from applied exploitation of the knowledge on the fungal siderophore system, the challenge of the future will be the elucidation of several unexplored aspects, e.g., details of the siderophore biosynthetic pathway, the mechanism of siderophore excretion, the mode of iron release from intracellular siderophores and the interplay of iron metabolism, reactive oxygen species (oxidative state) and fungal development, alone or in interactions with the host.
References:
Eichhorn H, Lessing F, Winterberg B, Schirawski J, Kamper J, et al. (2006). A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18:3332–45 Marcus Miethke and Mohamed A. Marahiel (2007). Siderophore-Based Iron Acquisition and Pathogen Control. MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS 71:413-451 Oide, S., Moeder, W., Krasnoff, S., Gibson, D., Haas, H., Yoshioka, K. and Turgeon B.G. (2006). (NPS6), encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18: 2836–2853. Oide S, Krasnoff SB, Gibson DM, Turgeon BG. (2007). Intracellular siderophores are essential for ascomycete sexual development in heterothallic Cochliobolus heterostrophus and homothallic Gibberella zeae. Eukaryot. Cell 6:1339–53 Schrettl, M., Bignell, E., Kragl, C., Joechl, C., Rogers, T., Arst, H.N., Jr, et al. (2004) Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med 200: 1213–1219. Schrettl, M., Bignell, E., Kragl, C., Sabiha, Y., Loss, O., Eisendle, M., et al. (2007) Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog 3: 1195–1207.
About Author / Additional Info:
Scientist, Plant Quarantine Division, ICAR- National Bureau of Plant Genetic Resources, Pusa Campus, New Delhi- 110 012