In recent years, numerous genome-sequencing projects identified large numbers of candidate open reading frames, to be annotated with a function. Analysis of these genes typically involves the transfer of DNA segments into a variety of vector backgrounds for protein expression and functional analysis. Often the cloning process involves restriction enzyme digestions. These digest set limits to the cloning step.
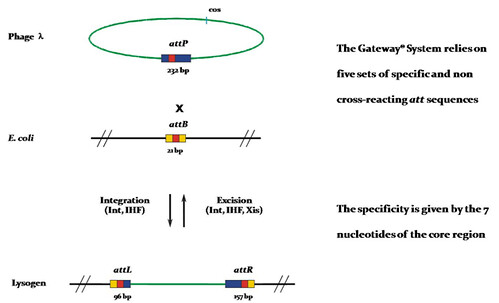
The Gateway Cloning System, invented and commercialized by Invitrogen since 2000 (Hartley et. al., 2000), is a molecular biology method that enables researchers to efficiently transfer DNA-fragments between plasmids. In E. coli bacterium, there is a stretch of DNA called attB and in the lambda phage there is a stretch of DNA called attP. After the phage infection, the recombination of injected lambda DNA with the corresponding bacterial DNA via the att sites in the presence of integration-specific enzymes (Int, IHF & Xis) takes place resulting in the integration of the phage DNA into the bacterial genome. Now, the hybrid recombination sites are called attL and attR (L stands for left, R stands for right) and these recombination reactions (“LR” and “BP”) are the basis of the Gateway Cloning System.
It provides an approach to high-throughput and in-depth functional analysis of genes for rapid optimization of protein expression by transferring numerous DNA segments parallel into many different vector backgrounds. The amino- and carboxy-terminal translation fusions can be generated, as the subclones maintains orientation and reading frame.
The main benefit of the Gateway system is that DNA sequence has to clone only one time to create an entry clone. Once the entry clone is verified, DNA fragment could moved across any expression system with a one-step, 1 hour recombination reaction (LR), without the use of any standard cloning reactions and screening of multiple clones.
The MultiSite Gateway technology, represent an extension of the Gateway site-specific recombinational cloning system, which can be utilize further for cloning of multiple DNA fragments in the same recombination cloning technique.
The Gateway cloning provides expression platform of choice, with a wide range of vectors for different plant systems and purposes (Earley et. al.,2006; Karimi et. al.,2007). Adapted Gateway vectors for Agrobacterium-mediated transformation, silencing studies, and many other applications have been already cited in over 150 peer-reviewed articles.
The basic cloning workflow involves isolation of RNA or DNA from various potential sources, having the gene custom isolated, or buying the gene of interest from a clone collection. The acquired gene is then usually cloned into an expression vector to perform functional analysis. Sometimes a restriction enzyme cannot be used because of the cut site within the gene of interest, which can truncate the gene to be cloned.
Gateway cloning eliminates the disadvantages of restriction enzyme based cloning and offers expression possibilities that have been impractical or involve many cumbersome steps with traditional restriction enzyme cloning. Manipulating large numbers of Genes was not possible in a uniform manner�"independent of size, sequence, or restriction sites. Gateway Cloning is nearly independent of these constraints and is highly efficient, so genes may be cloned, subcloned and then screened for phenotypes with high-throughput procedures using this technology.
References:
1. James L. Hartley, Gary F. Temple, and Michael A. Brasch (2000). DNA Cloning Using In Vitro Site-Specific Recombination. Genome Res. Vol. 10: 1788-1795.
2. Keith W. Earley, Jeremy R. Haag, Olga Pontes, Kristen Opper, Tom Juehne, Keming Song and Craig S. Pikaard (2006).Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal, 45, 616�"629.
3. Lee S-K, Kim B-G, Kwon T-R, Jeong M-J, Park S-R, Lee J-W, Byun M-O, Kwon H-B, Matthews BF, Hong C-B and Park S-C (2011). Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.). J. Biosci. 36, 139�"151.
4. Mansour Karimi, Ann Depicker, and Pierre Hilson(2007). Recombinational Cloning with Plant Gateway Vectors. Plant Physiology, Vol. 145, pp. 1144�"1154.
5. Manu J Dubin, Chris Bowler and Giovanna Benvenuto (2008). A modified Gateway cloning strategy for overexpressing tagged proteins in plants. Plant Methods, 4:3 doi:10.1186/1746-4811-4-3.
6. http://www.invitrogen.com/gateway
7. http://www.invitrogen.com/site/us/en/home/Products-and Services/Applications/Cloning/cloning-methods/plant-dna-cloning.html
About Author / Additional Info:
I am currently working as Assistant Professor at Amity Institute of Biotechnology, Amity University Haryana. I have obtained doctorate degree for development of Transgenic Bt chickpea plants in Biotechnology and Molecular Biology from CCS Haryana Agricultural University, Hisar as a CSIR/UGC- JRF(NET) fellow after completing Master’s in Biotechnology from DAV, College, PU, Chandigarh.