1. Mutation:
It is sudden heritable change in the gene sequence. When mutation occurs they can alter the gene product, prevent the gene from functioning or there would not be any effect at all. Without mutation all genes would exist in only one form. Alleles would not exist & a classical genetic analysis would not be possible. Most importantly populations of environment would not be able to evolve & adapt to new environments. Mutations could happen in gametic as well as somatic cells. Delicious apple and Novel orange are the examples of somatic cell mutations. Whereas, in gametic cell mutation; if they are dominant, then the effect is expressed immediately; on the other hand, if they are recessive, then effects are obscured in diploids.
Molecular basis of mutation:
a. Base substitution: Transition and Transversion
In this only one codon get substituted therefore only one amino acid get altered
b. Base addition or deletion:
If it is multiple of 3 then Reading frame do not change
If it is not multiple of 3 then frame-shift mutation occurs
c. Transposition: it occurs through Transposable elements. e.g. alleles producing wrinkled seed of pea
d. Trinucleotide repeat expansion: fragile X syndrome in human. In this syndrome, CGG repeat in FMR gene site of X chromosome becomes increasingly high. In normal individual, FMR site has 6 to 50 CGG repeats, whereas mutant FMR site has up to 1000 CGG repeats.
2. Transposable elements (TE):
These are Fragments of DNA which can move from one location to another in the genome. For millions of year, they are moving, copying and reinserting themselves at will. They can inactivate the gene in which they reside, cause chromosome breaks and transpose to new locations within the genome. TE can be classified in to two broad classes:
• Autonomous TE: They have ability to transpose e.g. Ac elements in maize
• Non-autonomous TE: they can transpose only with the help of autonomous elements e.g. Ds elements in maize
Transposable Elements in Prokaryotes:
a. Short sequences called Insertion Sequence (IS) elements: only possess genes required for transposes activity
b. Longer sequences called transposons: possess transposes activity + other genes
a. Bacterial Insertion Sequences: Are segments of Bacterial DNA that can move from one position on a chromosome to a different position on the same chromosome or on a different chromosome. All IS elements encode a protein, called a transposase, which is an enzyme required for their movement. In addition, they begin and end with short inverted repeat sequences that are also required for their movement. When IS elements resides in the middle of genes, they interrupt the coding sequence and inactivate the expression of that gene. IS elements can also block the expression of other genes in the same operon if those genes are downstream of the insertion site. E.g. IS1 is the 800-bp segment identified in gal.
b. Bacterial Transposons: There are two types of bacterial transposons
• Composite transposons: contain a various bacterial genes that reside between two nearly identical inverted IS elements (Figure1). Transposase encoded by one of the two IS elements is necessary to for movement of the entire transposon.
• Simple transposons: consist of bacterial genes flanked by short (<50bp) inverted repeat sequences. These sequences do not encode the transposase enzyme. Instead, simple transposons encode their own transposase in the region between the inverted repeat sequences. E.g. Tn3 ( Figure 1)
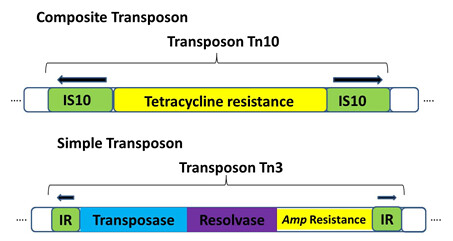
Figure 1: structure of composite and simple transposon
Mechanism of transposition: the activity of transposase can be divided in to two events Excision from the original location and Inserting into the new location.
Excision from the original location: almost all TE in prokaryotes and eukaryotes follow one of the two modes of transposition viz., replicative and conservative transposition. In replicative mode, a new copy of the TE is generated and gets inserted into the new site. The mechanism is similar to copy and paste. Whereas, in conservative mode, TE get excised from original location and get inserted in to new site which resembles cut and paste mechanism (Figure 2)
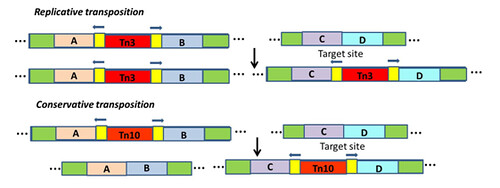
Figure 2. Mechanism of transposition of TE
Inserting into the new location: The transposase makes a five-base-pair staggered cut in the DNA. The TE inserts in between the staggered ends, and the host DNA repair machinery fills in the gap. At the site of insertion a five base pair direct repeat is formed, these sequences are called target-site duplication. All TE have target site duplication but its length could be vary according to type of TE.
Transposable Elements in Eukaryotes:
TE in Eukaryotes are of two classes: class 1 retrotransposons and class 2 DNA transposons
Class 1 retrotransposons: Are TE that employ enzyme Reverse Transcriptase to transpose through an RNA intermediate. They resemble animal viruses called retroviruses. Retrotransposon such as Copia in Drosophila (figure 3) has long terminal repeats at their ends are called LTR-retrotransposons. They use replicative mechanism to transpose.
Class 2 DNA Transposons: These transposons can insert in the genome by replicative or conservative mechanism. Elements that transpose in this manner are called class 2 elements or DNA transposons. First DNA transposon characterized is P element in Drosophila with a size of 2.9 kb and 31bp-Inverted repeat, it encodes a single proteinâ€"the transposase that is responsible for its movement (Figure 3)
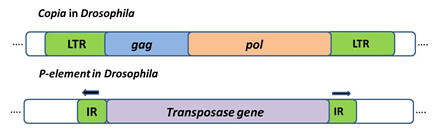
Figure 3: Transposable elements in eukaryotes
Transposable elements in Human Genome: more than half of the DNA in the human genome is likely derived from transposons. TE in humans are of two types; long interspersed elements (LINEs) and short interspersed elements (SINEs). LINEs do their movement with the help of reverse transcriptase (RT) but SINEs do not, as they do not encode their RT, rather they do their movement by RT encoded by LINEs. The most abundant SINE is Alu. One million whole and partial Alu sequences are present in human genome and make up >10% of human genome. Many LINEs, SINEs, Alu are found in the human gene (HGO) encoding homogentisate 1,2-dioxygenase, the enzyme whose deficiency causes alkaptonuria. If almost half of the human genome is derived from transposable elements then how do humans survive so many insertions in genes? The most likely answer is that, most of these TEs are located in intronic regions of genes.
3. Gene Duplication:
The first mechanism of new gene generation to be suggested is gene duplication (Ohno 1970). This is the most common way of creating new gene, with 20 times more frequency than mutation. Its classification is based on size and portion of the gene that is duplicated, entire genome may get duplicated, a large segment of a gene or individual gene or individual exon or specific part of a exon may get duplicated.
Mechanism of gene duplication:
i. Ectopic recombination: Duplication occurs due to unequal crossing over between misaligned homologous chromosomes. Sometimes crossing over also occurs at non homologous chromosomes. It do not require high level of homology between sequences, just 2.2kb of homologous stretch of nucleotides is enough. TE offers source of homologous repetitive DNA that can facilitate recombination (Zhang J 2003).
ii. Replication slippage: Errors in DNA replication produces duplication of short genomic sequence. During replication, DNA polymerase starts copying the DNA, at some point DNA polymerase dissociates from DNA and replication stalls. When the polymerase reattaches to DNA strand, it misaligns and copies of the same segment more than once (Hartl and Ruvolo 2012).
iii. Retrotransposition: if a replicating retrovirus invades a cell, its RT can attach cellular mRNA and retrogenes could form. These retrogenes may get inserted in the genome and it leads to changes in gene regulation.
iv. Aneuploidy: it occurs when nondisjunction of a single chromosome results in an abnormal number of chromosomes. Trisomy for Chr. 21 leads to Down syndrome in humans.
v. Polyploidy: Nondisjunction during meiosis results in additional copies of the entire genome. It is common in plants but also occurred in animals.
Evolutionary significance of gene duplication:
i. Neofunctionalization: in this event, original gene retains its function whereas duplicated gene- make new function through accumulation of mutations. This mutation is having no deleterious effect on the organisms. If original genes lost its function then the duplicated gene continue to function correctly (Lynch 2007; Conant and Wolfe 2008)
ii. Subfunctionalization: in this event, both copies are equally free to accumulate degenerative mutations as long as defects are complemented by the other copy. The original gene function is distributed among the two copies. No gene is lost but new function is not achieved (Force et al 1999).
4. Pseudogenes:
Pseudogenes are the molecular remains of broken genes, which are unable to function because of lethal injury to their structures. These are the sequences that resemble the known genes but cannot produce functional proteins. Pseudogenes originate through the same mechanism as protein coding genes followed by subsequent accumulation of disabling mutations (nucleotide insertion, deletion, substitution) that disrupt the reading frame or lead to insertion of a premature stop codon (Mighell et al 2000).
Pseudogenes can be classified in to,
Nonprocessed pseudogenes: usually contains introns, and are often located next to their parent genes.
Processed pseudogenes: originate through retrotranspositions, they lack introns, promoter region but often contains Poly A signal.
5. C value enigma:
Genome size does not correlate with the organism complexity e.g. Single cell amoebae have up to 100 fold larger genome than the humans. The genome contains substantial fraction of DNA other than functional genes e.g. The maize genome expanded by 50% in 140,000 years since its divergence from Zea luxuriance (Tenaillon et al 2011).
6. Gene amplification:
Gene amplified in large number 100-200 times of the original number. During oogenesis in many organisms a specific replication of the genes that code for ribosomal RNA occurs. During the development of Xenopus oocytes there is a special DNA synthesis that leads to a thousand fold amplification of the genes (900 copies) that code for ribosomal RNA (Hourcade et al 1973).
7. DNA Methylation:
It’s a biochemical process of adding methyl group to the cytosine by DNA methyltransferase. DNA methylation suppresses the expression of endogenous retroviral genes and other harmful stretches of DNA that were incorporated in the genome of the host overtime. Generally methylation occurs in a CpG sites. CpG rich regions in genome are called CpG islands. In mammals, CpG islands are associated with start of the gene (promoter region), which leads to silencing of the gene. In case of cancer, tumor suppressing gene (TSG) is silenced due to methylation (Craig and Wong 2011). In plants DNA methylation occurs at CpG, CpHpG and CpHpH sites where H represents any nucleotide but guanine. Plants are highly methylated eukaryotes with up to 50% of their cytosine exhibiting methylation. Interestingly, in fungi only repetitive DNA is methylated or in some species it is totally absent (Selker et al 2003).
8. Exon shuffling:
Eukaryotic genes consist of exons and introns. Exon shuffling is a process through which two or more exons from different gene can be brought together ectopically.
Mechanism of exon shuffling:
a. Crossover during sexual recombination of parental genome:
Eukaryotes are evolved by sexual recombination of parental genome.
i. Intron mediated recombination:
Generally introns are longer than exons and most of the crossing over occurs in non-coding regions. In these introns, large no of transposable elements repeated sequences promotes recombination of non-homologous genes which results in exon shuffling (figure 4).
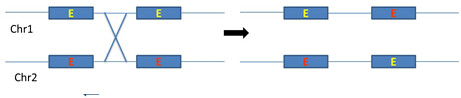
Figure 4: schematic representation of intron mediated recombination
ii. Unequal crossing over between exons of same chromosomes: Unequal crossing over between exons of same chromosomes produce real exon shuffling (figure 5)
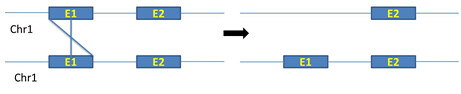
Figure 5: Unequal crossing over between exons of same chromosomes
b. Transposon mediated exon shuffling: occurs through
• LINE mediated exon shuffling: The most abundant LINE element in human is LINE1 (L1). LINEs replicate by copy â€"paste manner, when L1, mobilizes it gets associates with 3’ flanking DNA and carries the non-L1 sequence to a new genomic location location (Ejima & Yang 2003).
• LTR transposons mediated exon shuffling:
• Illegitimate recombination: is the recombination between short homologous sequences or nonhomologous sequences (Muszewska at al 2011).
9. Repetitive elements:
These are small fragments 500bp-1000bp sequence which is repeated in the genome several times. On the basis of no of repetitive sequence it can be Highly repetitive DNA >105 copies;
Moderately repetitive DNA 10-100000 copies and Less repetitive DNA 2-10 copies.
Repetitive sequence facilitate unequal crossing over because the repetitive sequence gives homology of DNA for crossing over to occur (figure 6)
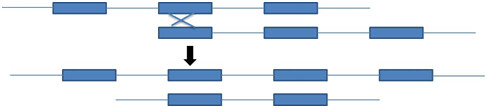
Figure 6: Unequal crossing over in the one generation leads to the change in the genome in the next generation
10. Horizontal gene transfer (HGT):
Its transfer of genes between organisms in a manner other than traditional reproduction. During bacterial evolution, the ability of Bacteria to adapt to new environments most often results from the acquisition of new genes through horizontal transfer rather than by the alteration of gene functions through numerous point mutations. Horizontal gene transfer is the primary reason for bacterial antibiotic resistance (Kay et al 2002). HGT has been an imp factor in the evolution of many organisms
Mechanism of HGT:
• Transformation : common in bacteria
• Transduction: through phage particles
• Conjugation: cell to cell contact
• Gene transfer agents: virus like elements encoded by host that are found in alphaproteobacteria
HGT in eukaryotes:
• DNA sequence suggest that HGT occurred within eukaryotes from chloroplast and mitochondria ( CHL and MT are probably bacterial endosymbiont of eukaryotic cell) (Blanchard and Lynch 2000)
• Striga has undergone HGT from Sorghum to its nuclear genome (Yoshida et al 2010)
• Genome of Pea aphid contains multiple genes that e=were horizontally transferred from fungi (Nancy et al 2010).
11. Chromosomal aberration :
It is well known that structural and numerical chromosomal aberrations are the important source of genetic variation in the population. Deletion, Duplication, Inversion, and Translocation and aneuploidy in the chromosome are the drivers of cellular evolution.
References:
1. Conant GC and Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics 9 (12): 938â€"950. doi:10.1038/nrg2482. PMID 19015656
2. Craig JM and Wong, NC (2011) Epigenetics: A Reference Manual. Caister Academic Press.
3. Ejima Y and Yang L (2003) Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum Mol Genet, 12(11), 1321-1328.
4. Force A, Lynch M, Pickett FB, Amores A, Yan YL and Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 (4): 1531â€"1545
5. Hartl LD and Ruvolo M (2012) Genetic Analysis of Genes and Genomes, Jones & Bartlett Learning, Burlington, pg. 529
6. Hourcade D, DresslerD and Wolfson J (1973) The amplification of ribosomal RNA genes involves a rolling circle intermediate. Proceedings of the National Academy of Sciences, 70(10), 2926-2930.
7. Kay E, Vogel TM, Bertolla F, Nalin R and Simonet P (2002) In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl. Environ. Microbiol. 68 (7): 3345â€"51.
8. Blanchard JL and Lynch M (2000) Organellar genes: why do they end up in the nucleus?. Trends Genet. 16 (7): 315â€"20.
9. Lynch VJ (2007) Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC evolutionary biology, 7(1), 2.
10. Mighell AJ, Smith NR, Robinson PA and Markham AF (2000) Vertebrate pseudogenes. FEBS Lett. 468 (2â€"3): 109â€"14.
11. Muszewska A, Hoffman-Sommer M and Grynberg M (2011) LTR retrotransposons in fungi. PLoS One, 6(12), e29425. doi: 10.1371/journal.pone.0029425 PONE-D-11-05922
12. Nancy A, Moran Tyler and Jarvik (2010) Lateral Transfer of Genes from Fungi Underlies Carotenoid Production in Aphids. Science 328 (5978): 624â€"627.
13. Ohno S (1970) Evolution by gene duplication. London: George Alien & Unwin Ltd. Berlin, Heidelberg and New York: Springer-Verlag..
14. Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP and Freitag M (2003) "The methylated component of the Neurospora crassa genome". Nature 422 (6934): 893â€"897.
15. Tenaillon MI, Hufford MB, Gaut BS and Ross-Ibarra J (2011) Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome biology and evolution, 3, 219-229.
16. The dynamic genome: Transposable elements
17. Yoshida Satoko, Maruyama Shinichiro, Nozaki Hisayoshi and Shirasu Ken (2010) Horizontal gene transfer by the parasitic plant Striga hermonthica. Science 328 (5982): 1128.
18. Zhang J (2003) Evolution by gene duplication: an update. Trends in Ecology & Evolution 18 (6): 292â€"8.
About Author / Additional Info:
Scientist at Division of Genetics, Indian Agricultural Research Institute. Working on Genetic Improvement of Wheat.