Author: Waghmare Sandesh Tulshiram
Sandeshwaghmare174@gmail.com
Article Summary: RNA editing is the post-transcriptional alteration of RNA sequences through insertion, deletion or substitution of nucleotides, excluding the RNA processing events. The gene whose transcript is edited is cryptogene and the addition of several Uracils during the insertion is termed as pan editing. Substitution includes both Cytosine (C) to Uracil (U) and Adenosine (A) to Inosine (I), through hydrolytic deamination. RNA editing could be used for genome manipulation in crop plants also.
1. Introduction
RNA editing is the post-transcriptional alteration of RNA sequences through insertion, deletion or substitution of nucleotides, excluding the processing events such as G-capping, polyadenylation and splicing. This process is often necessary to generate large number proteins with limited coding genes. RNA editing is a molecular process in which cells can make discrete changes to specific nucleotide sequencewithin a RNA molecule. Changes in nucleotide sequence has generated by RNA Polymerase. RNA editing has been observed intRNA,rRNA, mRNA or miRNA molecules of eukaryotes. RNA editing occurs in the nucleus and cytosol of mammals, as well as mitochondria and plastids of plant cell.
RNA editing provides a suitable platform in which a single gene is able to code a number of proteins. As a result, molecular diversity arise which finally reflect on phenotype and its leads to evolution. Evolution is defined as successive processes that bring changes in organisms to adopt a particular environmental condition. Evolutionary changes occur due to natural selection, mutation and genetic drift.
2 Types of RNA editing
RNA editing is mainly consists of four types like 1) Insertion of Uracil (U), 1) Deletion of Uracil (U) 3) Insertion of Cytosine (C), 4) Substitution of Cytosine to Uracil (C to U) and 5) Substitution Adenosine (A) to Inosine (I).
2.1. Uracil Insertion: This type of RNA editing is commonly observed in mitochondrial transcripts of Kinetoplastid protozoa. In pan editing several uridyl residues are incorporated. This process is guided by a small 3’- oligo-uridylated RNA (Guide RNA) and a set of enzyme complex called editosome. Editosome is also known as 20S enzyme complex. It consists of a series of enzymes which includes, Endonuclease, Terminal Uridyl Transferase (TUTase), 3’- Uridine exonuclease (3’-U-exo), RNA ligase.

Fig. 1: Pan editing
Guide RNA is a Small RNA molecules that are encoded by the genes present in the minicircles of mitochondria. Minicircle and maxicircle are two structures present in kinetoplast of mitochondria of Trypanosome; contain both non-coding and coding genes respectively. The guide RNAs are encoded by non- coding genes. The 5’ end contains anchor sequences which are complementary to target RNAs and 3’ end have a ten numbers of uridyl residues. Guide RNAs hybridized with the target RNA and it recruits the editosome.
Mechanism of Uracil Insertion- Uracil insertion involves four steps.
i) Hybridization of guide RNA with target RNA
ii) Removal of mismatches
iii) Incorporation of uridyl residues
iv) Sealing of edited RN
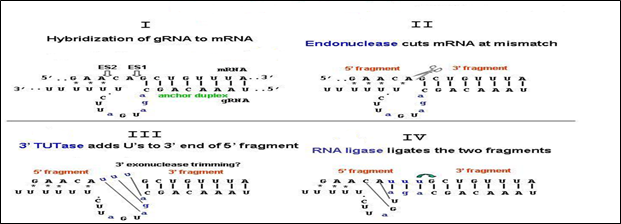
Fig 2: Mechanism of Uracil insertion
2.2. Uracil deletion: - Uracil deletion use similar enzymatic tools as Uracil insertion.
Mechanism of Uracil deletion- Uracil deletion involves four steps.
i) Hybridization of guide RNA with target RNA
ii) Removal of mismatches
iii) Uracil exonuclease which excises the (U)s at the deletion site
iv) Sealing of edited RNA
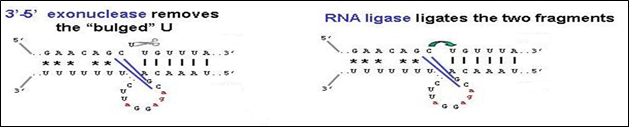
Fig 4: Mechanism of Uracil deletion
2.3. Cytosine insertion: This type of RNA editing was first observed in a slime mold Physarium polycephalum. Cytosine is incorporated in pre-mRNA which overcomes the frame shift after every 25 nucleotides.
2.4. Cytosine to Uracil substitution: Cytosine to Uracil substitution is the most common type of RNA editing observed in mitochondria and chloroplast of higher plants and some cases nucleo-cytoplasm of mammals. The process involves is hydrolytic deamination of cytosine which replace it to uracil. It is catalyzed by the enzyme cytosine deaminase.
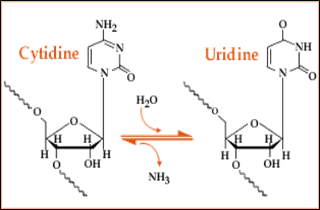
Fig. 5: Hydrolytic deamination of Cytosine where NH2 reacts with water, ammonia is released and finally Uracil is produced .
2.5 Adenosine to Inosine substitution:
Adenosine to Inosine substitution is reported in mammals but no such report is observed in plants. It is reported only in nuclear encoded RNAs. The multi-domain enzymes ADARs (Adenosine Deaminases Acting on RNAs) are involved in hydrolytic deamination of Adenosine which converts it to Inosine. The ADARs are classified in to three types based on the RNA they are acting and also presence and absence of functional domains.
ADARs enzyme
ADARs contain a highly conserved catalytic deaminase domain (DM) at their C-terminal Crystallography structure of the DM showed that the surface of this domain contains a positively-charged cleft for the binding of negatively-charged dsRNA and that it catalyses the hydrolytic deamination of Adenosine via a catalytic zinc ion. Moreover, an inositol hexakisphosphate (IP6) was found buried within the enzyme core that contributes to the protein fold. A nucleotide “flip-out” mechanism is necessary to force the targeted Adenosine into the catalytic pocket in the correct orientation for the deamination reaction.
The ADAR enzymes contain a C-terminal conserved catalytic deaminase domain (DM), two or three dsRBDs in the N-terminal portion. ADAR1 full-length protein also contains a N-terminal Zα domain with a nuclear export signal (NES) and a Zβ domain, while ADAR3 has a R-domain. A nuclear localization signal is also indicated.
3. Proteins essential for initiation of RNA editing
Two protein families play significant role in initiating the process of RNA editing viz.,
- Pentatrico Peptide Repeats (PPRs)
- Multiple Organellar RNA editing factors (MORFs) 3.1. Pentatrico Peptide Repeats (PPRs)
3.2. Multiple RNA Editing Factors (MORFs)
MORFs are proteins; provide additional components of RNA editing machinery in both plant organelles. Two MORF proteins are required for editing in plastids and at least two are essentials for editing in mitochondria. The loss of a MORF protein abolishes or lowers editing at multiple sites, many of which are addressed individually by PPR proteins. The morf gene belongs to a small family of nine genes and one potential pseudogenes. Of the encoded proteins MORF 2 and 9 are reported to be essential for chloroplast RNA editing and remaining MORF 3-8 are for editing of mitochondrial transcripts.
4.1. RNA editing in retrotransposons
Retrotransposons are the transposable elements which require and RNA intermediate for their transformation from one region to another region of same or different chromosome. Here Adenosine to Inosine RNA editing is observed. It is reported that the due to A to I editing for a period of time the intronic segments are converted into exonic segments and the process is called exonization. This is a mechanism which generates phenotypic diversity and also ability to create new function.
5. Applications of RNA editing
MtDNA is well understood to be a candidate to assess the evolutionary process and phylogeny. The mtDNA and cpDNA bound RNA editing and the accumulation of sequence variation is proportional to the evolutionary process. Understanding on the sequence variations brought in by this process throws light on the evolutionary process in a species. Human nuclear prelamin A recognition factor contains a primate specific Alu-exon that exclusively depends on RNA editing for its exonization. Abundant RNA editing of Alu sequences is the mechanism behind the birth of new exons in the human genome.
RNA editing could be used for genome manipulation in crop plants also. RNA editing has role in regulation of genes for chlorophyll biosynthesis, fertility restoration and enhancement of plant immunity
Conclusion:
As RNA editing is a natural process it helps a particular gene to express in a various way by generating novel Open Reading Frames (ORFs), its change the function of a particular cell when require. RNA editing could be used for genome manipulation in crop plants also. The role of RNA editing in regulation of genes for chlorophyll biosynthesis fertility restoration and enhancement of plant immunity are well understood.
References:
- Maor, G.L., Sorek, R., Levanon, E.Y., Paz, N., Eisenberg, E., and Ast, G. 2007. RNA-editing mediated exon evolution. Genome Biol. 8: R29.
- Takenaka, M., Zehrmann, A., Verbitskiy, D., Kugelmann, M., Härtel, B., and Brennicke, A. 2012. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. USA 109: 5104-5109.
- Yagi, Y., Tachikawa, M., Noguchi, H., Satoh, S., Obokata, J., and Nakamura, T. 2013. Pentatricopeptide repeats involved in plant organellar RNA editing. RNA Biol. 10: 1419-1425.
About Author / Additional Info:
I am currently working as Assistant professor at K.K. Wagh College of Agricultural Biotechnology, Nashik