Authors: Basavaprabhu L. Patil*, Subodh Kumar Sinha, S. V. Amitha Mithra and Amolkumar U. Solanke
ICAR-National Research Centre on Plant Biotechnology, IARI, Pusa, New Delhi
*Corresponding author: blaptil2046@gmail.com
Segmented RNA viruses are widely distributed in nature and are known to infect plants, animals and humans. Viral RNA segmentation helps in their reassortment during co-infections of different viruses. Thus, the progeny of such a reassorted virus contain genes derived from more than one parent resulting in fitness advantage or disadvantage. However, for survival and proliferation of reassorted viral progeny, the compatibility in their packaging signals and the continued maintenance of RNA and protein interactions is critical for their packaging in one single virion. Members of the order Bunyavirales which are made up of multiple, linear, single-stranded, negative sense or ambisense RNA segments are known to exhibit reassortment. The genus Emaravirus is one of the newest plant infecting genera of Bunyavirales, which is included in a newly created family Fimoviridae. Pigeonpea sterility mosaic emaraviruses (PPSMV) which are widely present in India are known to cause Sterility mosaic disease (SMD) of pigeonpea. There are two different species of PPSMV involved in SMD of pigeonpea, which are called asPigeonpea sterility mosaic emaravirus-1 (PPSMV-1) and Pigeonpea sterility mosaic emaravirus-2 (PPSMV-2). Recent studies on variability of PPSMVs by Patil et al. (Viruses 2017, 9, 183; doi: 10.3390/v9070183) has revealed wide spread occurrence and mixed infection of PPSMV-1 and PPSMV-2 across India and are also shown to exhibit viral segment reassortment. Such a phenomenon may complement for genetic drift and have major implication on evolution of PPSMVs in India. Although more than 300 viral species are reported to show segment reassortment, this is the first report (Patil et al., 2017) for an Emaravirus.
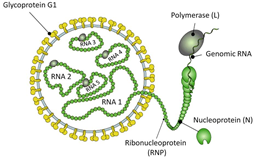
Figure 1: Conceptual diagram of PPSMV particle structure (source: Viral Zone, Swiss Institute of Bioinformatics; modified from Patil& Kumar, 2015).
References:
1. McDonald, S.M., Nelson, M.I., Turner, P.E., Patton, J.T. (2016) Reassortment in segmented RNA viruses: Mechanisms and outcomes. Nat. Rev. Microbiol. 14, 448-460.
2. Patil, B.L., Dangwal, M., Mishra, M. (2017) Variability of emaravirus species associated with sterility mosaic disease of pigeonpea in India provides evidence of segment reassortment. Viruses 9(7). pii: E183. doi: 10.3390/v9070183.
3. Patil, B.L., Kumar, P.L. (2015) Pigeonpea sterility mosaic virus: a legume-infecting Emaravirus from South Asia. Mol. Plant Pathol. 16, 775-786.
4. Patil, B.L., Kumar, P.L. (2017) Pigeonpea sterility mosaic emaravirus: a journey from a mysterious disease to a classic emaravirus. In: A Century of Plant Virology in India. Editors: B Mandal, GP Rao, VK Baranwal & RK Jain, Springer. Chapter 10, 255-270.
About Author / Additional Info: